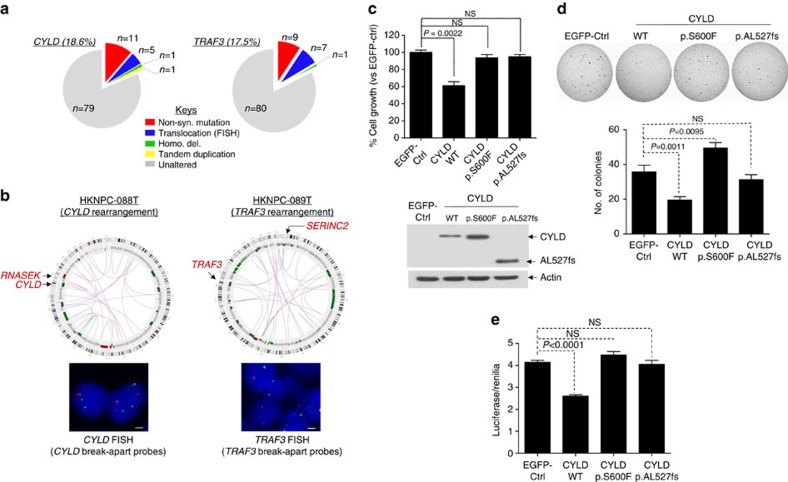
Figure 5. CYLD and TRAF3 alterations in NPC.
(a) Additional genetic alterations of CYLD and TRAF3 in NPC detected by fluorescent in-situ hybridization (FISH). Left: Five additional cases of CYLD rearrangements and 1 case of CYLD homozygous deletion were identified by FISH using a CYLD break-apart probe and 1 case of CYLD tandem repeat was confirmed by Sanger sequencing (Supplementary Figs 7 and 8). Right: Seven additional cases of TRAF3 gene rearrangements were also identified using TRAF3 break-apart probes. (b) Left: CYLD gene rearrangement as revealed by WGS. A circos plot showing the CYLD gene rearrangement (HKNPC-088T, confirmed by FISH). A scale bar representing 1 μm is shown in all FISH pictures. Right: TRAF3 gene rearrangement as revealed by WGS. A circos plot showing the TRAF3 gene rearrangement (HKNPC-089T, confirmed by FISH). (c) Transient transfection of CYLD wild-type (WT) gene into an EBV-positive NPC cell line, C666-1 resulted in significant growth inhibition at day 5 (7 × 104 cells in 4% FBS, P=0.0022, n=6). Similar results were obtained in three independent experiments. Expression of CYLD mutants (p.S600F and frameshift mutant p.AL527fs) revealed a loss-of-function of the tumour suppressive activity versus CYLD WT gene in C666-1 cells. CYLD WT and mutant protein expressions are shown underneath. (d) CYLD WT expression, but not the mutants, inhibited the anchorage-independent growth ability of C666-1 cells in soft-agar colony formation assay. C666-1 cells were infected with retroviral vectors expressing the CYLD WT gene and the CYLD mutants. Colonies were stained with 0.1% indonitrotetrazolium chloride and counted, and presented as a bar graph, which showed cumulative results of five independent experiments (n=15). (e) CYLD WT expression was able to reduce NF-κB activity in C666-1 cells in complete culture medium containing 10% FBS. Expression of CYLD mutants was not able to suppress NF-κB activity in C666-1 cells. The NF-κB activity was measured using the luciferase/Renilia assay (Promega, USA) with the Cignal NF-κB Pathway Reporter gene system (Qiagen, USA). A cumulative plot of five independent experiments (total n=19) showing changes in NF-κB-dependent luciferase activity in CYLD stable versus EGFP-Ctrl cells.