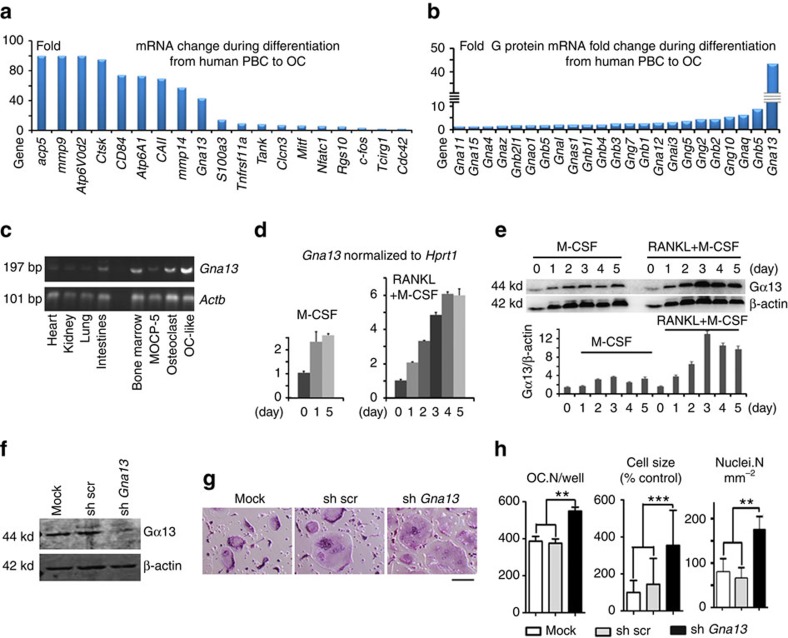
Figure 1. Gα13 which is induced by RANKL and M-CSF inhibits osteoclastogenesis.
(a,b) Microarray profile of gene expression during osteoclast differentiation. OC, osteoclasts; PBM, peripheral blood monocytes. (c) Semi-quantitative reverse transcription–PCR to detect Gna13 mRNA level in murine tissues and cells. (d) Quantitative reverse transcription–PCR to analyse time-course expression of Gna13, normalized to Hprt1, in murine bone marrow monocytes (BMMs) under the treatment of M-CSF, or M-CSF and RANKL; N≥4. (e) Western blot to analyse time-course expression of Gα13 in BMMs under the treatment of M-CSF or M-CSF and RANKL, and its quantification in lower panel; N≥4. (f) Western blot to analyse Gα13 expression in non-infected BMMs (mock) and BMMs infected with lentivirus expressing scrambled short hairpin RNA (shRNA) (sh-src) or shRNA targeting Gna13 (sh-Gna13). (g) TRAP staining to detect osteoclast formation of mock, sh-src and sh-Gna13 BMMs. (h) Quantification of cell number (OC.N/well and nuclei.N mm−2; N≥10) and relative cell size (N≥100) in g. OC.N/well, osteoclast (TRAP positive multinucleated cell) number per well. Nuclei.N mm−2, osteoclast nuclei number per mm2. Results in d,e,h are expressed as mean±s.d.; **P≤0.01; ***P≤0.001 (Student's t-test). Scale bars, 200 μm.