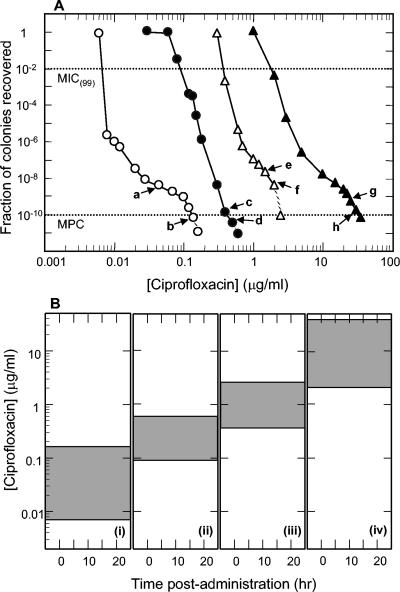
FIG. 1.
Stepwise enrichment of ciprofloxacin-resistant H. influenzae mutants. (A) Effect of ciprofloxacin concentration on recovery of colonies. Wild-type H. influenzae ATCC 49247 (open circles) was applied to ciprofloxacin-containing agar at the indicated concentrations, and colonies were recovered. Arrows indicate ciprofloxacin concentrations used to obtain colonies for which nucleotide sequence information was obtained. Arrows a and b indicate recovery of Ser-84-to-Tyr GyrA variants. Strain KD2308 (arrow b) was used for second-round selection (solid circles). Cells from positions c and d contained an additional parC mutation that changed Ser-84 to Arg. Strain KD2322 (arrow d) was used for a third round (open triangles). Cells from positions e and f contained an additional gyrA mutation that changed Asp-88 to Asn. Strain KD2364 (arrow f) was used for a fourth round (solid triangles). Cells from positions g and h contained an additional parC mutation that changed Glu-88 to Lys. Dashed lines, drug concentrations at which no colony was recovered; dotted lines, guides for determining MIC(99) and MPC, as indicated. (B) Relationship of mutant selection window to serum drug concentration. Shaded areas represent mutant selection window defined by MIC(99) and MPC, which were obtained from data shown in panel A. (i) Wild-type cells; (ii) first-step mutant; (iii) second-step mutant; (iv) third-step mutant.