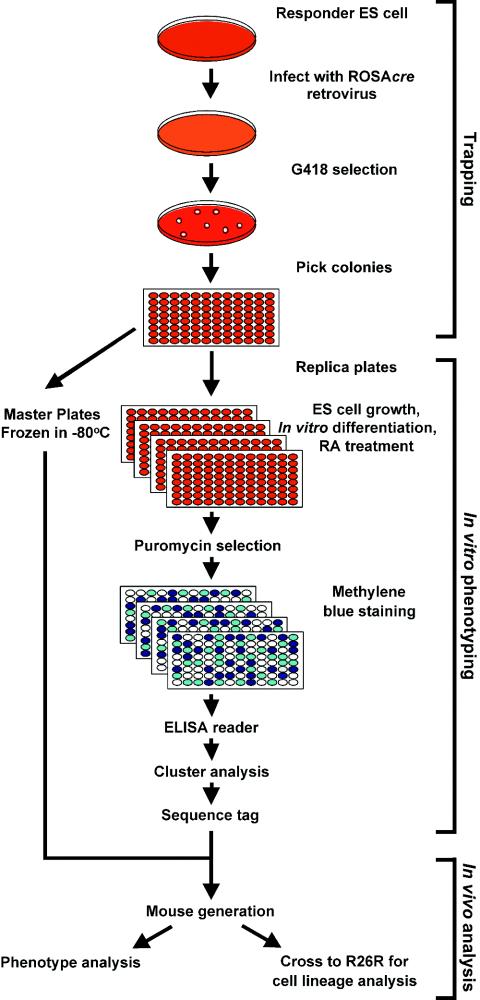
FIG. 3.
Flowchart illustrating the inducible gene trapping system to screen for gene traps of interest in vitro. Responder ES cells were infected with the ROSAcre trapping virus, followed by selection of trapping vector integration events in G418. G418-resistant colonies were picked and replicated, and a set of master plates were frozen at −80°C for future in vivo experiments. Replica plates were treated with different inducing conditions or exposed to various signaling molecules. After the treatment, puromycin selection was applied to each clone. The number of cells surviving puromycin selection represents the accumulated Cre-catalyzed drug resistance events induced by the conditions or signaling molecule. The absorbance readings at 600 nm of methylene blue-stained plates reflect the number of cells which survived drug selection, which is extrapolated to be an indicator of the expression level of the cre reporter. Multiple readings from different clones were used for cluster analysis. Gene trap clones from the subgroup of interest were used to obtain sequence tags with either 5′ RACE or inverse PCR. Clones of interest were used to generate mutant mice, allowing in vivo functional characterization of the trapped gene. ROSAcre-trapped mice can be crossed to R26R mice to trace the cell lineages of the trapped gene.