Figure 4.
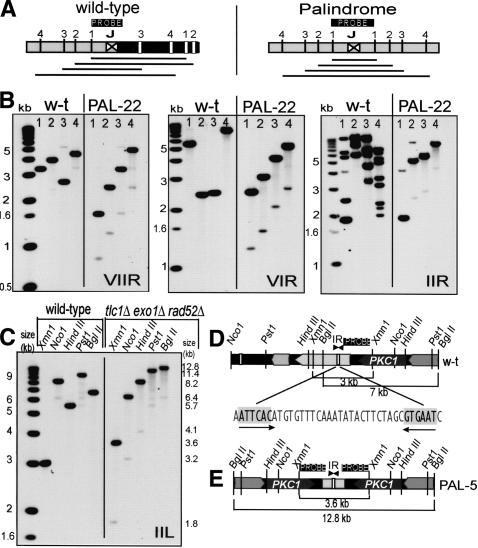
Palindromes form on chromosome arms. (A) Cartoons demonstrating how palindrome symmetry can be identified by appropriate choice of restriction enzymes and probe location. (Left cartoon) A succession of restriction enzymes (1, 2, 3, 4) will give a random pattern of bands when cutting wild-type DNA. (Right cartoon) If a palindrome has formed at this locus (by deleting the right half of wild-type DNA and duplicating the left half), enzymes will cut symmetrically around the junction between the palindrome arms, and increasing fragment sizes will generate a ladder. (B) Palindromes detected on the right arm of chromosomes VII, VI, and II in PAL-22. In each case, the first lane from the left shows molecular weight markers, the next four lanes are wild-type DNA, and the four right lanes are PAL-22 DNA (yku70Δ mre11Δ exo1Δ rad52Δ at 240 d). The enzymes used to cut DNA are indicated with numbers above each lane as follows: for chromosome VII, Stu1 (1), Bst11 (2), EcoR1 (3), and Bgl2 (4); for chromosome VI, Bst11 (1), Kpn1 (2), Pst1 (3), and Sac2 (4); and for chromosome II, Nsi1 (1), Afl2 (2), EcoR5 (3), and Nsp1 (4). The junction loci are shown in Figure 6. In the case of chromosome IIR, the probe hybridized close to the end of chromosome II, but also to several other wild-type chromosome ends, which were lost in this PAL-survivor by 240 d. Primers used to make probes are listed in Supplementary List 1. (C) A palindrome formed near PKC1 on the left arm of chromosome II. The first lane from the left shows molecular weight markers, the next five lanes are wild-type DNA, the five right lanes are DNA from PAL-5 (tlc1Δ exo1Δ rad52Δ strain at 100 d). The enzymes used to cut DNA and their positions are shown in D and E. (D) A map of the wild-type locus examined in C. Restriction enzymes sites, probe location, open reading frames, and an inverted repeat are shown. (E) A map of the palindrome detected in C.