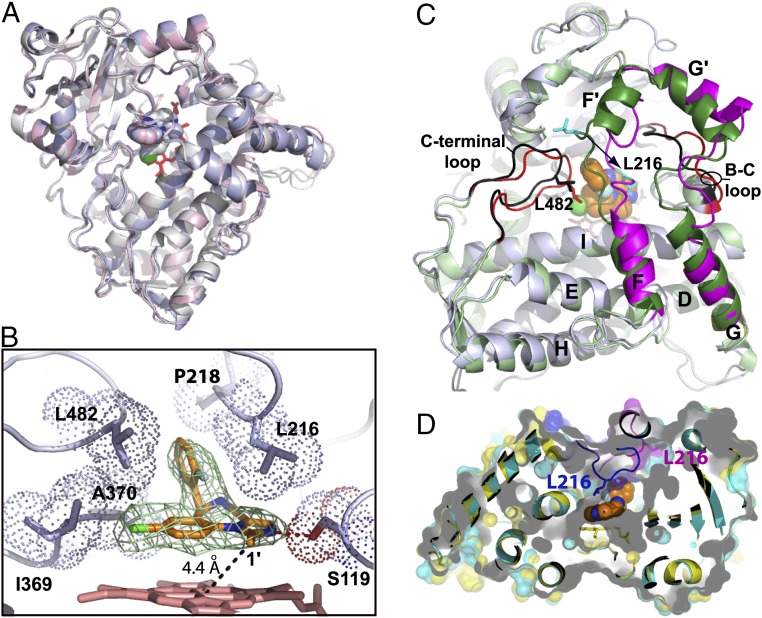
Fig. 3.
Crystal structure of the CYP3A4–MDZ complex. (A) Superposition of three molecules present in the asymmetric unit shows their similar fold (root-mean-square deviation between the ordered Cα-atoms is 0.6–0.7 Å). (B) Close-up view at the active site of molecule A. MDZ is bound above the heme plane with the C1–Fe distance of 4.4 Å and forms a hydrogen bond with Ser119 via the imidazole ring nitrogen. The chlorophenyl ring is within the Van der Waals distance from the Ile369–Ala370 fragment, whereas the fluorophenyl ring is flanked by the Leu216, Pro218, and Leu482 side chains. These interactions immobilize MDZ and orient suitably for hydroxylation of the C1-atom, the primary oxidation site. Simulated annealing Fo–Fc composite omit map for MDZ (in green mesh) is contoured at 3σ. (C) Superposition of the ligand-free (Protein Data Bank ID 1TQN; in light/dark green and black) and MDZ-bound (in gray, magenta and red) structures. Structural elements undergoing reorganization are labeled. MDZ is in orange and space-filling representation. A 12-Å shift of Leu216 (shown in cyan) is indicated by an arrow. (D) Slice through the superimposed 1TQN (cyan and magenta) and MDZ-bound structures (yellow and blue) with semitransparent surfaces to show that the substrate-induced conformational switch leads to the active site collapse.