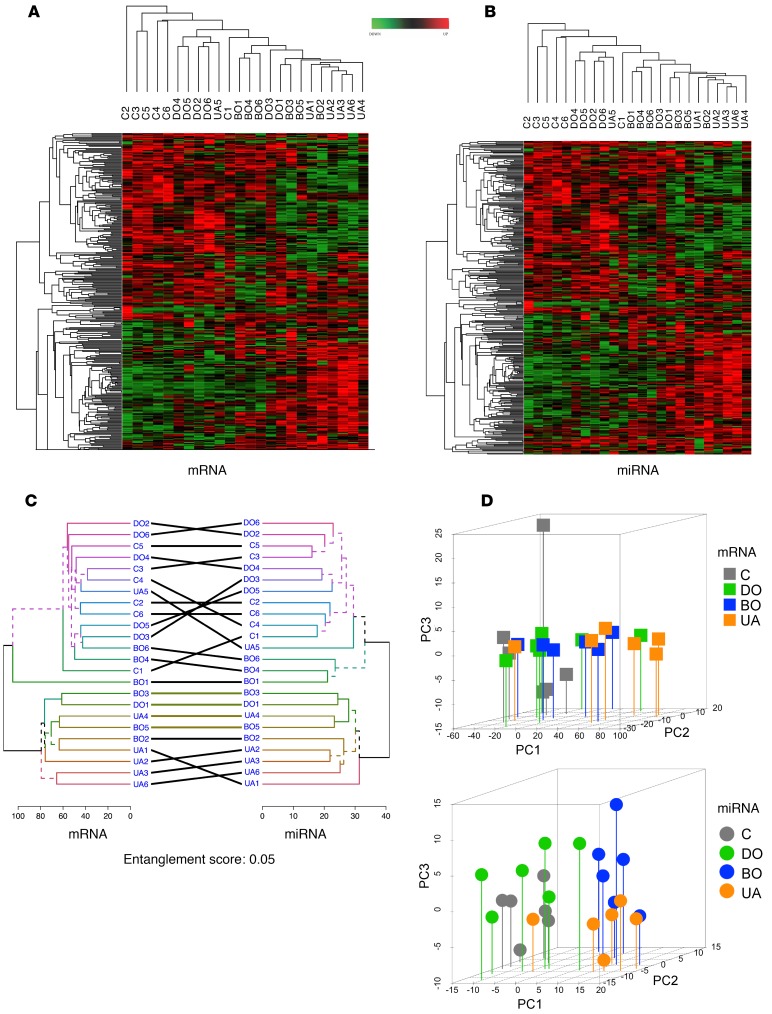
Figure 2. Differentially expressed mRNAs and miRNAs in BOO-induced LUTD patients.
(A) Hierarchical clustering and heatmap of log2 fold change of 2,634 significantly regulated mRNAs (y axis) in 24 patients (x axis). (B) Hierarchical clustering of 343 significantly regulated miRNAs (y axis) and 24 samples (x axis). Hierarchical clustering of mRNAs and miRNAs revealed high expression profile similarity between DO and controls, whereas BO is similar to UA. (C) miRNA-mRNA tanglegram. Unique nodes, with a combination of patients not present in the other tree, are highlighted with dashed lines. Entanglement has the score of 0.05, indicating a very high similarity between the dendrograms of both mRNA and miRNA samples. (D) Principal component analysis of mRNA and miRNA expression was done based on profiles of 10,064 mRNAs (top) and 343 miRNAs (bottom) for each group (6 patients per group). The position of patients from each group on 3D plots indicates the similarity of mRNA or miRNA expression patterns. PCA confirms the sample clustering shown in the heatmaps, with controls clustered with DO and BO clustered with UA. The intergroup dispersity is low, indicating sample homogeneity. C, controls; DO, obstructed patients with detrusor overactivity; BO, obstructed patients without detrusor overactivity; UA, obstructed patients with underactive bladders.