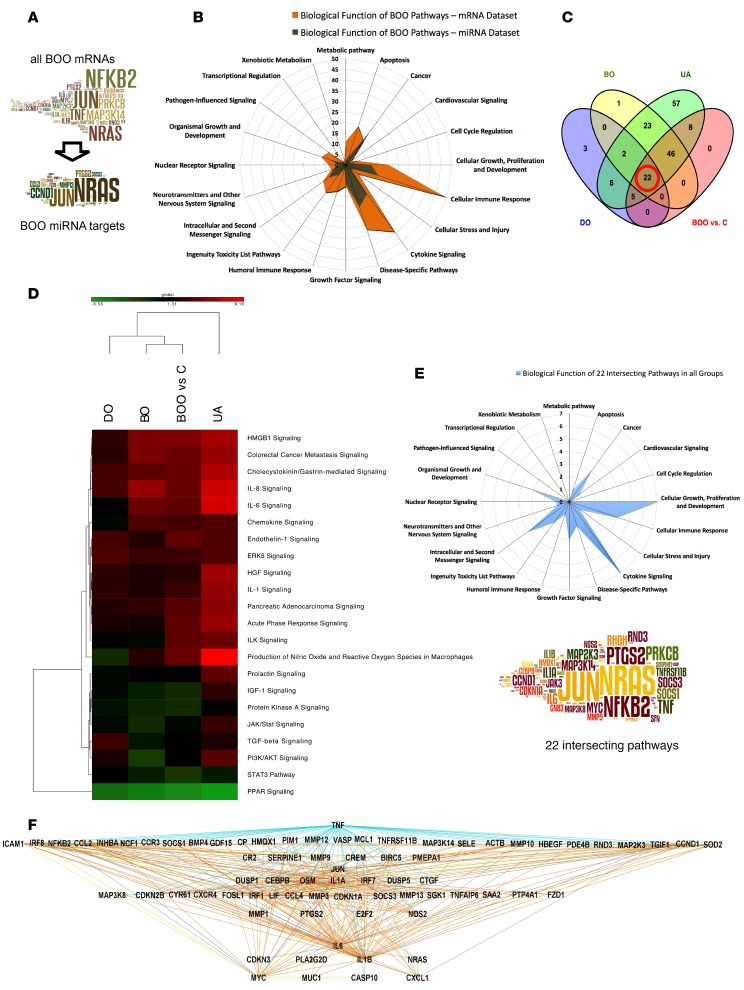
Figure 8. Pathway analysis in all BOO patients and identification of 22 key signaling pathways.
We investigated mRNAs and miRNAs differentially expressed in all bladder outlet obstruction (BOO) patients (n = 18) compared with controls (n = 6) and performed gene enrichment analysis using mRNAs and the expressed targets of 12 abundant miRNAs. (A) The word clouds show frequent pathway elements in the mRNA data set of all BOO patients and pathway elements targeted by abundant BOO miRNAs. Many signaling hubs in BOO are potentially miRNA regulated. (B) The radar graph for biological functions of significant (P < 0.05) BOO mRNA data set pathways (orange) and pathways built using expressed BOO miRNA targets (brown). The miRNA-controlled processes contribute to fewer biological functions. (C) Venn diagram of common regulated pathways (mRNA data sets, P < 0.05) in 3 patient groups (DO, BO, and UA) intersecting with the all BOO patient group; 22 common pathways are indicated. (D) Heatmap and clustering based on Z score of 22 common pathways. Z scores of the common pathways in all BOO patients resemble the scores in the BO group. (E) Radar graph for the biological functions of 22 common pathways of all BOO patients. The word cloud shows top pathway elements, and font size indicates the frequency of occurrence. Cellular growth, proliferation and development, and cytokine signaling were the main biological functions shared by all groups. (F) Hierarchical view of upstream analysis mechanistic network made of transcription factors and cytokines in 22 common pathways (mRNAs) of all BOO patients. TNF is the top upstream regulator, connected to 63 downstream elements, including NFkB, JUN, and MYC. C, controls; DO, obstructed patients with detrusor overactivity; BO, obstructed patients without detrusor overactivity; UA, obstructed patients with underactive bladders.