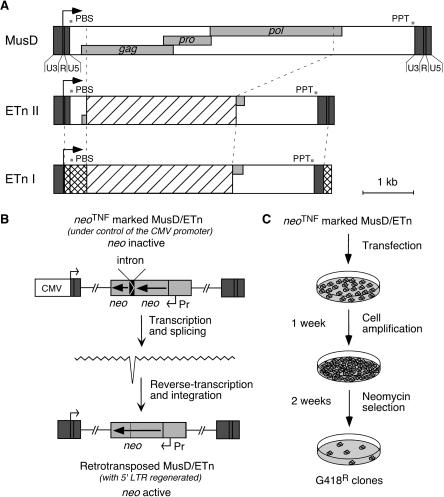
Figure 1.
Structure of the related MusD and ETn elements and rationale of the assay for retrotransposition. (A) Genomic organization of MusD, ETn type II and I elements. The LTRs (dark gray box) with a U3-R-U5 organization border three ORFs homologous to the retroviral gag, pro, and pol genes (light gray boxes). The transcription start site is marked with an arrow, and the signals necessary for retroviral reverse transcription are indicated: (PBS) primer binding site; (PPT) poly purine tract. ETn elements are closely related to MusD despite the replacement of the retroviral genes by a noncoding sequence (hatched box). ETn I elements differ from ETn II only in the LTRs and 5′-UTR region (squared boxes). (B) Schematic representation of a MusD/ETn element under the control of the CMV promoter and marked with the neoTNF reporter gene (light gray), in which the neo gene placed in backward orientation with its own promoter (Pr) is rendered inactive by the presence of a forward intron (top), which should be spliced out in the transposition RNA intermediate (middle), thus resulting in an active neo gene in the de novo MusD/ETn insertion (bottom). (C) Experimental procedure for detection of MusD/ETn retrotransposition. Cells were transfected either with a neoTNF-marked defective MusD/ETn together with an MusD expression vector, or with a full-length neoTNF-marked MusD alone. Cells were amplified for a week, and retrotransposition events were detected upon G418 selection.