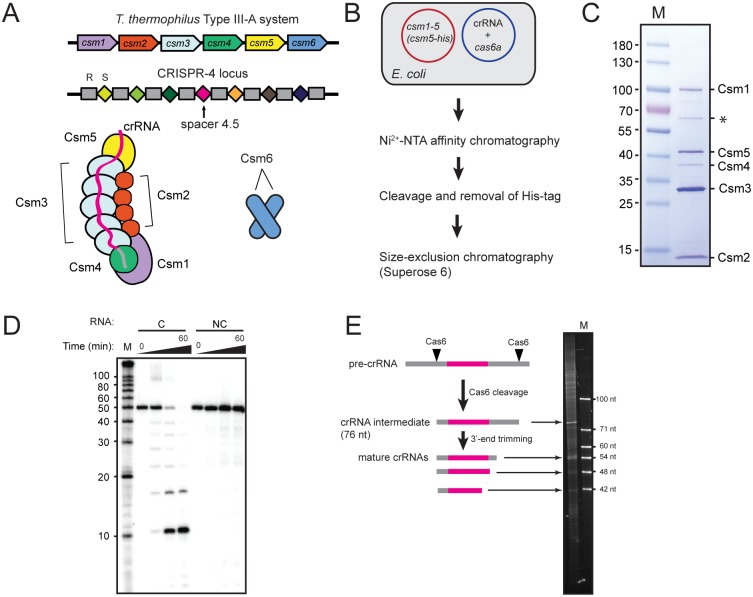
Fig 1. Reconstitution and RNA cleavage activity of a T. thermophilus Csm complex (TthCsm) purified from E. coli with a defined crRNA species.
(A) Components of the CRISPR locus and effector complexes of the T. thermophilus Type III-A Csm system. The complex is shown with 5 copies of Csm3 and 4 copies of Csm2, but complexes with different numbers of these two subunits also exist. The CRISPR-4 locus associated with the system is shown (repeat is designated by R and spacer by S). The spacer 4.5 used for complex reconstitution encodes for one of the most abundant crRNAs found in the host organism [21]. (B) Reconstitution and purification of TthCsm in E. coli. A plasmid containing genes encoding for Cas10/Csm1, and Csm2-5, with a His10 tag on Csm5, was co-transformed into E. coli with a plasmid containing genes for expression of T. thermophilus Cas6A and a single CRISPR array containing one copy of spacer 4.5. The purification steps are indicated. (C) TthCsm was subjected to SDS polyacrylamide gel electrophoresis (SDS-PAGE) analysis following purification. Csm subunits are labeled, and a molecular weight ladder (M) is in the left lane (masses are given in kilodaltons). A GroEL contaminant (asterisk) was also identified by mass spectrometry (S2 Table). (D) TthCsm-mediated cleavage of a complementary (C) or noncomplementary (NC) 32P-labeled ssRNA oligonucleotide was tested in the presence of 2 mM MgCl2. Samples taken at 0, 5, 30, and 60 minutes after TthCsm addition were analyzed by denaturing PAGE. (E) Schematic of crRNA processing in Type III CRISPR-Cas systems is shown on the left. Pre-crRNAs are cleaved by Cas6 to generate an intermediate, which is then trimmed at the 3’-end, resulting in mature crRNAs. On the right, nucleic acids associated with the Csm complex were extracted and analyzed by denaturing PAGE. An ssDNA oligonucleotide ladder (M) was loaded in the right-most lane and nucleotide lengths are indicated.