Abstract
Circadian clocks influence virtually all physiological processes, including lactation. Here, we investigate the role of the CLOCK gene in regulation of mammary epithelial cell growth and differentiation. Comparison of mammary morphology in late-pregnant wild-type and ClockΔ19 mice, showed that gland development was negatively impacted by genetic loss of a functional timing system. To understand whether these effects were due, in part, to loss of CLOCK function in the gland, the mouse mammary epithelial cell line, HC11, was transfected with short hairpin RNA that targeted Clock (shClock). Cells transfected with shClock expressed 70% less Clock mRNA than wild-type (WT) HC11 cultures, which resulted in significantly depressed levels of CLOCK protein (P < 0.05). HC11 lines carrying shClock had four-fold higher growth rates (P < 0.05), and the percentage of cells in G1 phase was significantly higher (90.1 ± 1.1% of shClock vs. 71.3 ± 3.6% of WT-HC11) following serum starvation. Quantitative-PCR (qPCR) analysis showed shClock had significant effects (P < 0.0001) on relative expression levels of Ccnd1, Wee1, and Tp63. qPCR analysis of the effect of shClock on Fasn and Cdh1 expression in undifferentiated cultures and cultures treated 96 h with dexamethasone, insulin, and prolactin (differentiated) found levels were reduced by twofold and threefold, respectively (P < 0.05), in shClock line relative to WT cultures. Abundance of CDH1 and TP63 proteins were significantly reduced in cultures transfected with shClock. These data support how CLOCK plays a role in regulation of epithelial cell growth and differentiation in the mammary gland.
Keywords: circadian, CLOCK, lactation, mammary development
dynamic changes occur in molecular clocks across multiple tissues during the transition from pregnancy to lactation. Amplitude and robustness of rhythms of clock genes in SCN and liver are increased. In the mammary gland, the stoichiometric relationship of core clock proteins change, with the ratio of BMAL1 and CLOCK increasing relative to a decrease in PER2 levels during the transition from pregnancy to lactation (5). In vitro studies demonstrated that these changes in mammary core clock dynamics were associated with development. Moreover, temporal expression patterns of core clock genes change throughout mammary development, and these changes were coincident with changes in peak proliferation (pregnancy) and differentiation (lactation) state of the gland (23). The significance of these changes and the role of molecular clocks, in particular, the mammary clock, in regulation of lactation competence are currently not understood.
Studies of circadian regulation of reproduction in the Clock-Δ19 line of mice found this mutation has minimal effects on growth and development of pups during gestation, but litter growth and survival are significantly decreased postnatally (7, 14, 15). The increased pup mortality rate is evident in both heterozygous and homozygous Clock-Δ19 mutant offspring; thus, increased pup mortality is likely due to a maternal defect in circadian clocks that causes a decrease in lactation competency in this line of mice. Clock-Δ19 mice have an N-ethyl-N-nitrosourea-induced mutation that affects transactivation properties of CLOCK and results in disruption of behavioral rhythmicity, loss of rhythmic gene expression, and downregulation of CLOCK-BMAL1 target genes (2, 16, 26). Thus, in the Clock-Δ19 mutant line, circadian clocks are compromised in both central and peripheral tissues. Dolatshad et al. (7) compared Clock-Δ19 to Vipr2−/− lines of mice to understand whether impact on litter growth and survival was due to loss of circadian clock function centrally vs. across the whole animal, as mice with a null mutation of the VPAC2 receptor gene (Vipr2−/−) only have a deficient clock in the master clock in the suprachiasmatic nuclei-SCN (13), while peripheral tissue molecular clocks are not directly affected. They found pups born to Clock-Δ19 dams had significantly reduced survival to weaning compared with pups born to wild-type or Vipr2−/− dams, suggesting that loss of peripheral clock function was impairing lactation competence in the Clock-Δ19 line. Lactation competency can be affected by multiple interacting factors that range from maternal behavior to systemic hormones to mammary development. Hoshino et al. (14) reported altered maternal nursing behavior in the Clock-Δ19 line of mice. Specifically, they found that the daily rhythm of maternal nursing behavior had a strong diurnal peak and two weak nocturnal peaks in wild-type dams, whereas Clock-Δ19 dams exhibited no significant peaks in activity. Moreover, the duration of nursing bouts was significantly longer in wild-type mice vs. Clock mutants. However, the number of nursing events per day was greater in Clock-Δ19 line vs. wild-type animals. Hoshino et al. (14) also found wild-type, but not Clock-Δ19, dams showed circadian rhythms of prolactin serum content; however there was no significant difference in mean daily prolactin levels between the lines. Thus, although maternal behavior and systemic hormones are impacted by Clock-Δ19 mutation, it is not likely that mutation effects on nursing behavior and prolactin can fully explain the poorer lactation competence in this line of mice.
Endogenous clocks generate circadian rhythms through a series of interlocked transcription-translation feedback loops. At the core of the loops are two transcription factors, CLOCK (or its paralog NPAS2) and BMAL1. CLOCK-BMAL1 heterodimers bind the E-box regulatory element in promoter regions of genes (12), including Period (Per1 and Per2) and Cryptochrome (Cry1 and Cry2) genes. PER and CRY proteins function as negative regulators of CLOCK-BMAL1-mediated transcription (1, 25, 31). The 24-h periodicity in activation-repression of the molecular clock results in circadian oscillation in ~10% of mammalian transcriptome, which translates into daily variations in cellular, metabolic, and physiological functions (25). Rhythmic output genes differ among tissues, allowing circadian control of function and activity appropriate for each organ. In the mammary gland, 7% of genes expressed in the gland during lactation exhibit circadian oscillation, and these genes function to regulate cell proliferation and differentiation, as well as metabolic output (20). Cell cycle regulators and tumor suppressors have been identified as clock-controlled genes in multiple tissues (9, 25, 28), and circadian control of these genes is believed to be important to the coupling of regulation of proliferation with key tissue functions. Thus, it is likely that circadian clocks play a similar role in the mammary gland. In addition, our previous studies of cattle showed that exposure to chronic shifts of light-dark cycle, which disrupts the circadian system, decreased milk production and impacted genes important to milk synthesis (4), suggesting that clocks play an important role in regulation of milk synthesis.
We hypothesize that the circadian clock in the mammary gland plays an integral role in regulating mammary gland development, as well as metabolic output during lactation, and, thus, mutation or decreased expression of the CLOCK gene in mammary epithelial cells will impact growth and differentiation. Here, we show that in late pregnancy, morphological development of the mammary gland was impaired in Clock-Δ19 mice compared with wild-type dams. Poorer development of the gland was associated with lower survival rates of pups born to Clock-Δ19 dams. To determine whether loss of CLOCK function in mammary epithelial cells could account for this phenotype, we used a mouse mammary epithelial cell line, HC11, to study the impact of decreasing CLOCK levels with short hairpin RNA (shRNA) on growth and differentiation of cultures. HC11 cells undergo differentiation upon lactogen treatment, and our previous studies demonstrated periparturient changes in mammary clock dynamics can be mimicked in culture upon treatment with prolactin and glucocorticoids (5). Here, we report that reducing CLOCK levels in HC11 cells, increased growth rates, and decreased expression of factors associated with mammary differentiation and metabolic output.
MATERIALS AND METHODS
Animal studies.
All animal work was performed in the Roswell Park Cancer Institute (RPCI) vivarium with Institutional Animal Care and Use Committee approval. C57BL6/J wild-type (WT) male and female mice were purchased from the Jackson Laboratories (Bar Harbor, ME) and were maintained on 12:12-h dark-light dark cycle with ad libitum access to food and water. The breeding colony of Clock-Δ19 mice was maintained in RPCI vivarium. Female WT and Clock-Δ19 mice were mated on a 1:1, female:male ratio with WT males, and checked daily for vaginal plugs (pregnancy day 1).
Mice were anesthetized with 5% isoflurane with 1 l/min O2 on pregnancy days 16 and 17 blood and abdominal mammary glands were collected at 6 and 10 h (n = 3/genotype/time point) after lights on. Blood was collected, and plasma prepared and stored at −20°C until analysis. Commercially available enzyme immunoassay kits were used to measure plasma prolactin (Calbiotech, Spring Valley, CA) and corticosterone (ALPCO, Salem, NH) levels, following the manufacturer's directions. Mammary number 4 glands were removed and fixed in 10% buffered formalin for histological analysis. The number of fetuses were counted and recorded.
Histological analysis of mammary morphology.
After overnight fixation in buffered formalin, mammary tissue was transferred to PBS. Tissues were paraffin-embedded, and 7-μm sections were cut and placed on electrostatically charged slides. Hematoxylin and eosin (H&E)-stained sections were used to evaluate the impact of ClockΔ19 mutation on mammary development. Slides were blinded by treatment, and to account for potential heterogeneity of tissue, three images of each H&E section were captured using a digital camera attached to a light microscope. The three areas were selected by first focusing scope at the approximate center of tissue section, and then the field of view was moved to 12, 4, and 8 o’clock relative to center, and the image was captured. Image Pro Plus software was used to measure luminal area and area of alveolar epithelium in three images with n = 6 animals/treatment. The ratio of luminal area to alveolar epithelium was calculated and was expressed as means ± SE.
Cell culture.
HC-11 cells were routinely cultured in complete growth medium [RPMI 1640 supplemented with 2 g/l sodium bicarbonate, 100 U/ml penicillin, 100 µg/ml streptomycin, 10% heat-inactivated FCS, 5 µg/ml insulin (Ins), and 10 ng/ml epidermal growth factor (EGF)] in 5% CO2 at 37°C. Cells were passaged 1:3 when they reached 80% confluence by harvesting cells with 0.25% trypsin EDTA.
Four unique Qiagen SureSilencing shRNA plasmid sequences specific to the mouse Clock gene (Table 1) were transfected into HC-11 cells. Each shRNA plasmid carried a U1 promoter, a shRNA sequence, and a hygromycin resistance gene. Plasmids were amplified in Escherichia coli and were isolated according to the manufacturer’s protocol (Endofree plasmid maxi kit; Qiagen, Germantown, MD) and were linearized with restriction enzymes (New England BioLabs, Ipswitch, MA). For transfection, HC-11 cells were plated at a density of 100,000 cells/ml in 24-well cell culture plates with complete growth media. Linearized Clock shRNA plasmids were transfected into HC-11 cells using the lipofection Attractene Transfection Reagent (Qiagen cat. no. 1051561). When transfected cells reached confluence, hygromycin was added at 200 µg/ml. Cells that were viable after 7 days of hygromycin selection were used to create clonal lines by dilutional cloning. Clonal cell lines were screened for level of Clock mRNA using quantitative PCR (qPCR). Nontransfected HC-11 cells served as wild-type control.
Table 1.
Nucleotide sequence of shRNA transfected into HC11 cells
| Sequence ID | Insert Sequence |
|---|---|
| shClock-1 | AAACCCACATTCCTTAGTAAT |
| shClock-2 | GCAACTTGTGACCAAATTAGT |
| shClock-3 | CGATGTCTCAAGCTGCAAATT |
| shClock-4 | ATCAAACCCTGGATTGAATTT |
| Negative control (scramble) | ggaatctcattcgatgcatac |
For growth curve analysis, 100,000 cells were plated using complete growth media in 6-well dishes, and cells from two dishes per time point were harvested using trypsin EDTA and counted on days 2, 4, and 7 using a Bio-Rad (Hercules, CA) TC10 automated cell counter. Growth curves were conducted four times, and results are presented as mean number of cells each day ± SE. Doubling time was calculated using the tool available here (33).
For cell cycle analysis, cells were plated overnight at a density of 100,000 cells/ml in RPMI media supplemented with Ins and 10% FCS. Media were removed, and cells were washed twice with PBS and serum starved in RPMI media + Ins. After 24 h in serum-free media, complete growth media [RPMI with 10% FCS, Ins, and EGF] was added (time 0), and cells were collected every 4 h over a 48-h period. At each time point, cells were collected for cell cycle and qPCR analysis of gene expression. To label cells with propidium iodide (PI) for flow analysis of cell cycle phase, cell pellets of 1.5 ×106 cells were resuspended in 100 µl of PBS and fixed with 280 µl of ice-cold 90% ethanol. Fixed cells were stored at −20°C until day of analysis. On the day of analysis, fixed cells were pelleted by centrifugation and resuspended in 1 ml PBS-0.5% BSA, pelleted again, resuspended in PBS-0.5% BSA + 100 U/ml RNAse (R6513 Sigma-Aldrich), and incubated for 15 min at 37°C. Five microliters of PI (500 μg/ml stock solution) were added, and cells were incubated 15 min at RT. PI-labeled cells were analyzed by FACS using a Beckman Coulter FC500 instrument. FACS analysis was repeated three times.
To induce HC11 differentiation, cells were plated at 100,000 cells/ml and grown to confluence. At confluence, cells were washed twice with PBS, and incubated for 48 h in RPMI media supplemented with 10% FCS and Ins. Undifferentiated cultures were collected for isolation of RNA and protein after 48 h. For differentiated cultures, media were changed to RPMI supplemented with 10% FCS, dexamethasone (0.1 μM), insulin (5 μg/ml), and prolactin (5 μg/ml) (DIP treated), and incubated for 96 h, with media change every 2 days. After 96 h in DIP, differentiated cultures were collected for RNA and protein isolation.
Protein isolation and Western blot analysis.
Protein lysates were isolated from cultures by pouring off media, washing cells two times with chilled PBS, and harvested using a scraper and 3 ml of cold PBS. Cells were pelleted by centrifugation, and cell pellets were lysed for 30 min on ice with 600 µl of cell extraction buffer (Invitrogen, supplemented with 1 mM of PMSF and 50 µl/ml of Protease Inhibitor Cocktail), with vortexing at 10-min intervals. Protein lysates were transferred to a microcentrifuge tube and centrifuged at 13,000 rpm for 10 min at 4°C. Protein was then aliquoted into microfuge tubes and stored at −80°C until further analysis. Protein concentration was measured using the Quick Start Bradford Protein Assay (Bio-Rad).
Fifty micrograms of protein was loaded per lane and electrophoresed on a 10% TGX precast SDS PAGE gel from Bio-Rad. Protein was transferred onto a polyvinylidene fluoride membranes for Western blot analysis. Membranes were probed for CLOCK, BMAL1, PER2, e-cadherin-CDH1, TP63, and β-actin proteins using Anti-CLOCK (AbCam, ab3517, dilution 1:5,000), anti-CDH1 (Cell Signaling, Danvers, MA; 24E10, dilution 1:1,000) anti-TP63 (Millipore, Billerica, MA; ABS552, dilution 1:7,500) and Anti-β-actin (AbCam ab8227, dilution 1:7,500) antibodies, respectively. The secondary antibody (ab97051) was then incubated on the blots at the dilution of 1/10,000. Membranes were washed with the detection reagent Clarity Western ECL Substrate (Bio-Rad). Blots were imaged using the ChemiDoc MP system (BioRad), and the relative band intensity was measured using ImageJ software. Relative protein abundance was determined by calculating ratio of target protein to β-actin and then normalized relative to WT HC11 cultures by dividing by ratio value. To accommodate multiple treatments, there were times two gels needed to be run. When this was done, gels were loaded, run, and transferred to membranes simultaneously, followed by membrane incubation with antibody probes in the same solutions, to allow for band intensities to be compared across blots. Data are expressed as means ± SE of relative ratios across at least three experiments.
RNA isolation and real-time quantitative PCR analysis.
RNA was collected from cells and isolated using Qiagen’s RNeasy kit, and quantity was measured using Nanodrop. Gene expression was analyzed using TaqMan one-step real-time quantitative PCR analysis (RT-qPCR; Life Technologies). Multiple genes (β-actin, β-microglobulin, and 18S) were analyzed to serve as a reference (housekeeping gene) for calculating relative expression using the 2−ΔΔCT method (19); 18S was chosen as the reference gene based on its levels staying steady across time and genotype. Commercially available (Life Technologies, Carlsbad, CA) mouse-specific TaqMan assays were used to measure the expression of Clock, Fasn, TP63, Wee1, Csn2, Cdh1, Ccnd1, and 18S.
To calculate relative expression, mean ΔCT of wild-type cultures across all time points for cell cycle analysis or mean ΔCT of undifferentiated cultures was used as a normalizer for relative expression. Except where noted, data are presented as mean log base twofold change ± SE of relative expression levels.
Data and statistical analysis.
Statistical analysis was performed using Minitab software. ANOVA was used to analyze whether shRNA sequences significantly impacted variables (relative expression, protein abundance, doubling time, and cell density) relative to wild-type HC-11 cultures. General linear model was used to determine the effect of treatment and time on cell number. A Tukey test was used for post hoc pairwise comparisons. Significance was considered at P ≤ 0.05.
RESULTS
Impact of Clock Δ19 mutation on mammary development in late pregnancy.
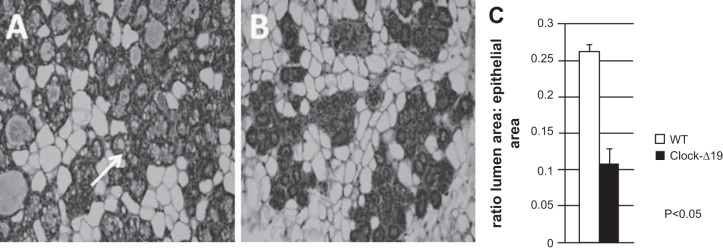
To explore the impact of loss of functional circadian timing system on mammary development, we compared morphology of glands from mutant Clock-Δ19 female mice with wild-type controls on day 16 of pregnancy. Consistent with previous reports (7), we found that although Clock-Δ19 females bred to wild-type (WT) males became pregnant and delivered offspring, dams had 0–4 pups present in the early postpartum period. Analysis of number of fetuses in utero on pregnancy day 16 showed no difference in litter size between WT and Clock-Δ19 mice (7.18 ± 0.95 and 6.88 ± 1.36, respectively), indicating that there was a high rate of neonatal mortality in litters of Clock-Δ19 dams. Using a milking machine designed for rodents (6), we were able to collect milk samples from Clock-Δ19 females on postpartum day 3. Although milk quantity was not sufficient for compositional analysis, visual inspection revealed a viscous yellow liquid, suggesting that gland differentiation was compromised. Compromised differentiation may be due to either failure to fully differentiate or involution changes in gland due to high loss of suckling neonates by day 3 in Clock-Δ19 line. Comparing mammary development between late-pregnant WT and Clock-Δ19 dams showed that the ratio of lumen to epithelial area in mammary tissue from WT dams was significantly greater in WT than Clock-Δ19 dams, thus indicating that deficiency in circadian clock impairs mammary development (Fig. 1). Basal plasma prolactin (mean across all time points) only tended to be different (P = 0.1) between WT (61.7 ± 17.6 ng/ml) and Clock-Δ19 (28.2 ± 9.3 ng/ml) mice on pregnancy day 16. There was no difference in basal corticosterone levels (98 ± 38.4 ng/ml and 86 ± 5.1 ng/ml, respectively). It is important to note that the larger standard error of mean basal plasma hormone levels of WT vs. Clock-Δ19 mice reflects temporal differences in WT animals vs. a lack of this difference in Clock-Δ19. These studies support that circadian clocks impact mammary development; however, we do not know whether these effects were due to loss of functional CLOCK in central, peripheral (and), or mammary clock.
Fig. 1.
Genetic knockout of functional circadian clocks negatively impacts mammary development in Clock-Δ19 mice. Hematoxylin-and-eosin (H&E)-stained 7-µm sections of mammary tissue collected on pregnancy day 16 from WT (n = 6) (A) and Clock-Δ19 (n = 6) (B) mice show that lack of functional circadian clocks impact mammary differentiation, including decreased formation of fat droplets (arrow) and lower ratio of lumen to total epithelial area (C), measured using image analysis software (Image ProPlus). Student's t-test analysis indicated ratio of lumen to epithelial was significantly different between WT and Clock-Δ19 mammary glands, P < 0.05.
Impact of decreased Clock expression on growth of HC11 cells.
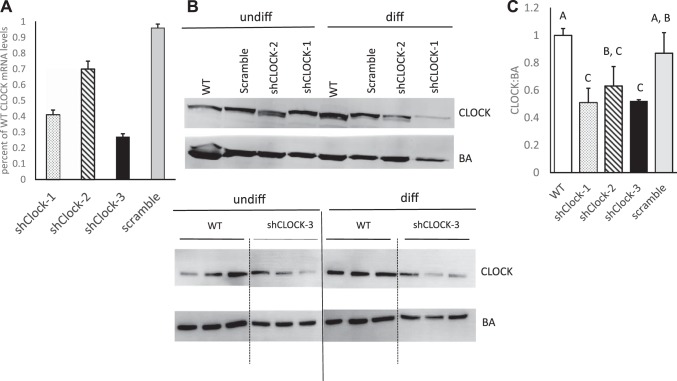
To determine whether CLOCK plays a role in regulation of mammary epithelial cell growth, HC11 cells were transfected with shRNA specific for CLOCK (shClock) or negative control scramble sequence (Table 1). Clonal HC11 cells carrying shClock sequence 3 (shClock-3) expressed Clock at 30% of control cultures (Fig. 2A), which resulted in significantly depressed levels of CLOCK protein (Fig. 2, B and C). HC11 cells carrying scramble shRNA sequence had no significant change in Clock mRNA or protein levels (Fig. 2).
Fig. 2.
Impact of shRNA sequence on CLOCK mRNA and protein levels relative to wild-type (WT) cultures. A: quantitative PCR (Q-PCR) analysis was used to determine the percent CLOCK mRNA levels in clonal cells lines established after transfection with shRNA sequences 1, 2, and 3 (shClock−1, −2, and −3, respectively) or negative control sequence (scramble) relative to WT HC11 cultures. B: representative Western blots of CLOCK and β-actin (BA) proteins; dashed-vertical lines indicate splicing of image to remove intervening lanes within same blot, solid-vertical lines indicate splicing between two different blots. C: densitometric analysis of Western blots and relative ratios of CLOCK:BA proteins across three experiments per clonal line. Different letters indicate difference at P < 0.05, as determined by ANOVA followed by a Tukey post hoc test.
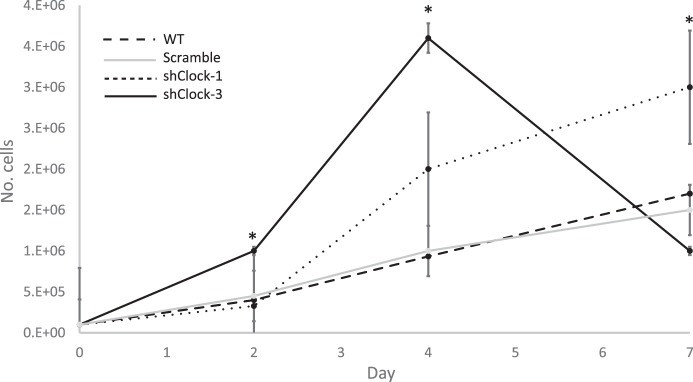
Seven-day growth curve analysis was used to determine the impact of shRNA sequence on HC11 growth. Growth pattern of cells carrying the negative control scramble sequence was not different from WT culture (Fig. 3), and doubling time between days 2 and 4 of culture was also not different (P > 0.05) between WT and scramble-transfected HC11 cells (36.1 ± 1.5 and 32.2 ± 0.1 h, respectively). However, transfection with shClock sequence 1 (shClock-1) or shClock-3 significantly reduced doubling times (23.5 ± 4.3 h or 18.5 ± 1.3 h, respectively) relative to WT cultures. The dramatic decline in cell number in shClock-3 cultures between days 4 and 7 was most likely due to overcrowding of cells.
Fig. 3.
Growth curve analysis of the impact of shRNA sequence on HC11 cells in culture. On day 0, 100,000 cells were plated in 35-mm dishes, and the number of cells were counted on days 2, 4, and 7 after plating. Two-way ANOVA of data from three experiments showed that day and line (WT: dashed line; shClock-1, dotted line; shClock-3, solid black line; scramble, solid gray line) had a significant effect (P < 0.05) on growth. The Tukey post hoc test indicated that cell numbers were different from control at P < 0.05 (*).
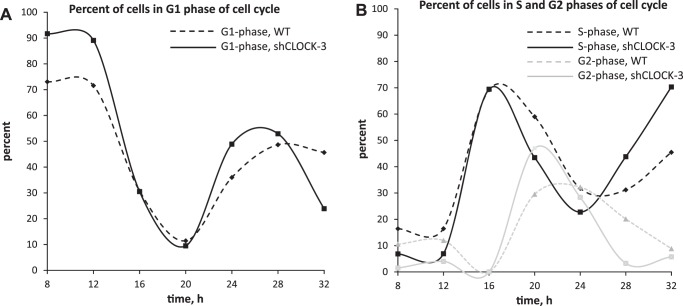
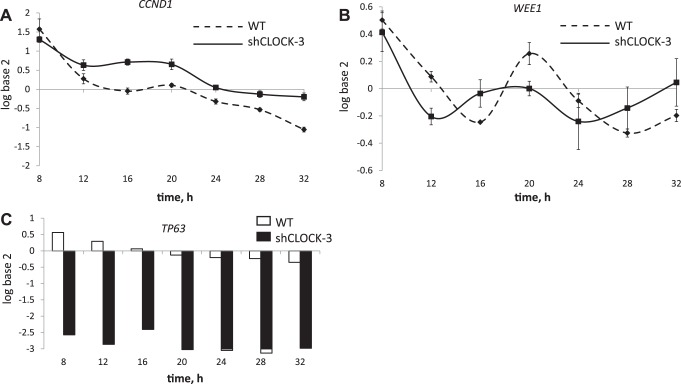
To investigate the consequence of reduced CLOCK abundance on progression through the cell cycle and cell cycle regulators, shClock-3 and HC11 cultures were plated, synchronized in the cell cycle by serum deprivation, released from arrest by the addition of serum plus EGF, and collected for FACS and q-PCR analysis. Patterns of cell cycle progression were highly similar among three replicate experiments, with graphs of one trial (Fig. 4) representing the 24-h window (8 h to 32 h) used to analyze mRNA expression of the cell cycle regulators CCND1, WEE1, and TP63. Cells transfected with shClock-3 appeared to be more sensitive to serum starvation, with 91.1 ± 2.1% of cells arrested in G1 phase of cell cycle at time 0, vs. 72.4 ± 13.3% in WT cultures (Fig. 4A). Consistent with a greater rate of growth was evidence for a more rapid succession from S to G2 phase, as there were closer and sharper peaks in these phases across the 24-h period in shClock-3 cultures compared with WT (Fig. 4B).
Fig. 4.
Cell-cycle analysis of the impact of shCLOCK on progression of HC11 cells through the cell cycle. WT (dashed) and shCLOCK-3 (solid) lines were seeded in 35-mm dishes and allowed to plate for 24 h in growth media. Cultures were serum starved for 24 h, and then growth media (10% serum + EGF) were replaced (time 0). Cultures were collected every 4 h over a 48-h period, and 1.5 million cells were fixed, stained with PI, and stage of cell cycle was determined using FACS. Graphs show difference in the percentage of WT and shCLOCK-3 lines in G1 phase (A) or S phase (black lines) and G2 phase (gray lines) (B) in a 24-h period, spanning from 8 h to 32 h, after addition of serum +EGF. Data are representative of one experiment; experiment was repeated three times.
Two-way ANOVA of temporal patterns of cell cycle regulator expression showed that time and shClock-3 significantly impacted CCND1 and WEE1 expression (P < 0.001). Steady-state levels of CCND1 were elevated in shClock-3 transfected vs. WT cultures across all time points, except during cell cycle arrest (8 h; Fig. 5A). WEE1 expression patterns were shifted, and amplitude decreased in cultures transfected with shClock-3 (Fig. 5B). Further, TP63 mRNA expression was significantly decreased (P < 0.05) by shClock-3 across all time points (Fig. 5C). We confirmed that effect of shClock-3 on cell cycle regulators was due to decreased CLOCK by measuring expression of CCND1 in shClock-1 and scramble lines. Similar to shClock-3, CCND1 levels were significantly elevated in shClock-1 lines, but CCND1 levels were not different between WT and scramble cultures (data not shown).
Fig. 5.
Impact of shCLOCK on expression of cell cycle regulators. WT and shClock-3 cells were plated in media with 10% serum, serum starved to synchronize cells, and then media with 10% serum + EGF were added (time 0). Cells were collected every 4 h over 48 h. RNA was isolated from cultures collected during the 8- to 32-h period, and Q-PCR analysis was used to measure steady-state CCND1 (A), WEE1 (B) and TP63 mRNA levels. Two-way ANOVA of data from three replicate experiments showed that time and line (WT vs. shClock-3) had significant (P < 0.001) impacts on CCND1 and WEE1 expression, and line had a significant (P < 0.001) impact of TP63 expression.
Impact of shClock on markers of mammary differentiation in HC11 cells.
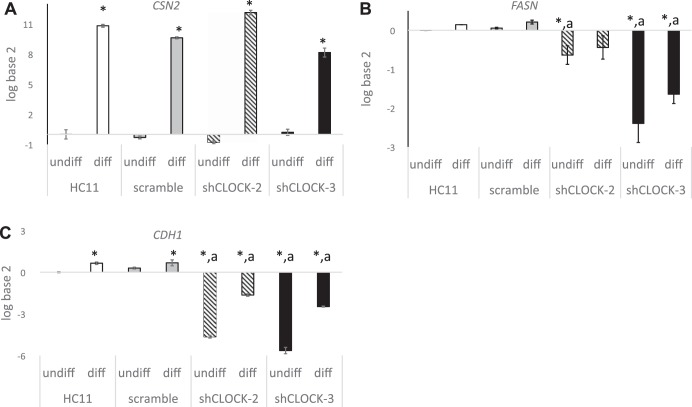
Our previous studies (4, 5, 30) and poorer survival rates of Clock-Δ19 litters suggest that circadian clocks may function to regulate differentiation and milk synthesis in the mammary gland. To determine whether this is regulated at the level of the mammary clock, we measured the impact of shClock on expression of e-cadherin (CDH1; Fig. 6), fatty acid synthase (FASN; Fig. 6), β-casein (CSN2; Fig. 6), and TP63 (Fig. 7) in undifferentiated (undiff) and differentiated (diff) HC11 cells.
Fig. 6.
Impact of shCLOCK on markers of differentiation in HC11 cells. WT, scramble, and shClock-3 cells were plated and grown to confluence in 10% + EGF, media were changed to 10% serum alone and undifferentiated (undiff) cultures were collected after 48 h. Differentiated (diff) cultures were incubated for an additional 96 h in media supplemented with dexamethasone, Ins, and prolactin + 10% serum. RNA was isolated, and qPCR analysis was used to measure steady-state CSN2 (A), FASN (B), and CDH1 (C), mRNA levels. Two-way ANOVA of three replicate experiments showed that state of differentiation had a significant impact on CDH1, FASN, and CSN2 expression and line (WT HC11, scramble, shClock-3) had significant (P < 0.05) effect on CDH1 and FASN expression. Tukey post hoc analysis demonstrated that expression level was different from HC11-undiff (*P < 0.05), or from HC11-diff (aP < 0.05).
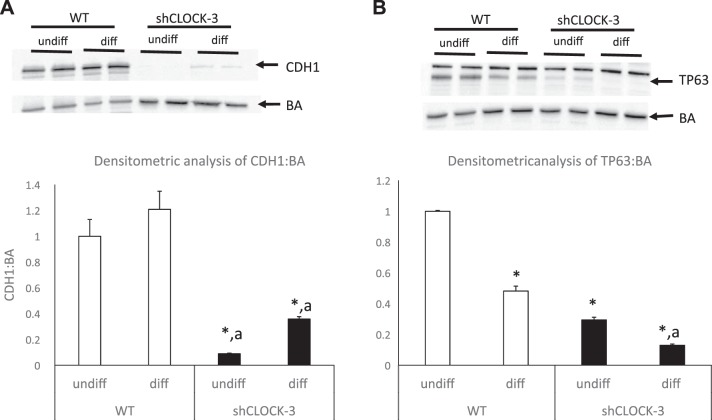
Fig. 7.
Impact of shClock on CDH1 and TP63 protein levels in undifferentiated (undiff) and differentiated (diff) HC11 cells. WT (open bar) and shClock-3 (solid bar) cells were plated and grown to confluence in 10% + EGF, media were changed to 10% serum alone, and undifferentiated (undiff) cultures were collected after 48 h. Differentiated (diff) cultures were incubated for an additional 96 h in media supplemented with dexamethasone, Ins, and prolactin + 10% serum. Protein lysates were prepared and used for Western blot analysis of CDH1 (A) and TP63 (B) levels. Two-way ANOVA of data from three experiments indicated that the state of differentiation and line had significant effects (P ≤ 0.05) on both CDH1 and TP63 levels. Tukey post hoc analysis demonstrated that expression level was different from HC11-undiff (*P < 0.05), or from HC11-diff (aP < 0.05).
CDH1 functions as an adherens junction protein, and its abundance is increased with differentiation of mammary epithelial cells (5). qPCR (Fig. 6) and Western blot (Fig. 7) analysis showed that CDH1 mRNA and protein levels were significantly decreased in shClock-3 transfected HC11 vs. WT cells. Cells transfected with shClock-3 also had significantly lower levels of Fasn relative to both undifferentiated and differentiated wild-type cultures; however, there was no effect of scramble sequence on steady-state levels of Fasn mRNA (Fig. 6). Levels of the gene that encodes the milk protein CSN2 were significantly increased in differentiated vs. undifferentiated states of HC11 cultures both in WT and shClock lines (Fig. 6). However, there was no difference in expression levels among wild-type, scramble, shClock-1 or shClock-3 transfected cultures.
Impact of shClock on core clock dynamics.
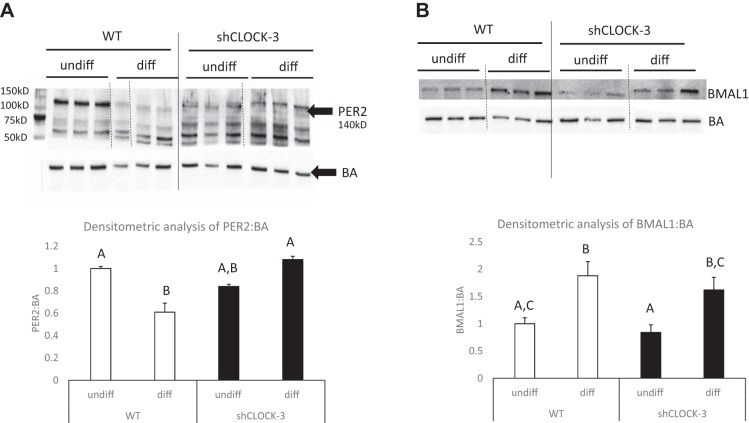
Our previous studies showed that HC11 cells have a robust clock (5). To determine the impact of shClock on other components of the core clock, cells were collected from undifferentiated and differentiated cultures. Similar to previous studies, we found that PER2 abundance was significantly less and BMAL1 protein abundance was significantly greater in cultures treated with lactogens for 96 h (diff) vs. undifferentiated wild-type HC11 cultures (undiff; Fig. 8); however, the abundance of PER2 was not different among undifferentiated and shClock-3 cultures treated with lactogens for 96 h (diff) (Fig. 8). In the shClock-3 lines, similar to wild-type HC11 cultures, BMAL1 abundance was greater in lactogen-treated cultures (diff) relative to undifferentiated shClock-3 treatment (Fig. 8).
Fig. 8.
Impact of shClock on PER2 and BMAL1 protein abundance in undifferentiated and differentiated HC11 cells. WT and shClock-3 cells were plated and grown to confluence in 10% + EGF, media were changed to 10% serum alone, and undifferentiated (undiff) cultures were collected after 48 h. Differentiated (diff) cultures were incubated for an additional 96 h in media supplemented with dexamethasone, Ins, and prolactin + 10% serum. Protein lysates were prepared and used for Western blot analysis of BA and PER2 (A) and BMAL1 (B) levels. Dashed vertical lines indicate splicing to remove intervening lanes within same blot, solid vertical lines indicate splicing between two different blots run simultaneously. Densitometric analysis was used to determine ratio of PER2 and BMAL1 to BA across three replicate experiments. Different letters indicate difference at P < 0.05, as determined by ANOVA followed by the Tukey post hoc test.
DISCUSSION
Our analysis of mammary morphology of Clock-Δ19 mice in late pregnancy showed poorer gland development. Poor pup survival and viscous-yellow mammary secretion of lactating dams also suggested mammary differentiation in Clock-Δ19 was negatively impacted. Others found that maternal behavior and serum prolactin levels are altered in Clock-Δ19 dams, however, these alterations are not likely to wholly cause the decreased lactation competency observed in this line (13, 14). Our studies with HC11 cell model system support that the impaired lactation competency observed in the Clock-Δ19 mice (7, 14, 15) may be due to lack of a functional clock in mammary epithelial cells.
Decreased CLOCK abundance resulted in increased mammary epithelial cell growth rate, decreased cell line doubling time, and a greater rate of progression through cell cycle. Cells transfected with shClock were also more responsive to serum starvation, suggesting that a fully functional clock may buffer against external change. This supposition is supported by studies that have shown tissue-specific rhythmicity can regulate the responsiveness of peripheral clocks to various stimuli, including temporal information from the SCN (24, 32). Changes in growth dynamics were accompanied by differential regulation of the cell cycle regulators WEE1, CCND1 and TP63. A strong correlation between the timing of clock gene expression with the timing of cell-cycle events has been demonstrated in regenerating liver (21) and continuously proliferating tissues (27). The coupling of circadian clock network with cell cycle network occurs through both direct and indirect control of the clock on cell cycle regulators. WEE1 is a direct transcriptional target of BMAL1-CLOCK (21). In our study, temporal pattern of WEE1 expression showed peak levels coincident with timing of transition from G1 to S and G2 to M in both wild-type and shClock-transfected cells, which is consistent with its cell cycle regulatory role (8). However, decreasing CLOCK levels in HC11 cells resulted in decreased WEE1 amplitude, supporting that known CLOCK-BMAL1 cell cycle regulatory targets were impacted by shClock transfection.
Previous studies have shown an association among circadian clock disturbances and changes in CCND1 expression. In Per2 mutant mice CCND1 temporal expression is altered (11), and osteoblasts isolated from PER1 and PER2 knockout mice exhibited shorter cell cycles and increased growth rates, which was accompanied by higher expression of CCND1 (8). In our study, we found that reduced CLOCK abundance in HC11 cells was associated with higher expression levels of CCND1, which is consistent with greater growth rates and increased rate of progression through cell cycle measured in shClock transfected lines.
Also consistent with greater growth rate and increased rate of progression through cell cycle in shClock-transfected lines was the dramatically reduced levels of TP63 mRNA and protein levels. TP63 encodes two main isoforms, TAp63 and ΔNp63, both of which are expressed in the mammary gland (10). TAp63 isoforms can bind to DNA through canonical p53-responsive elements and activate the transcription of many p53 target genes. As a result, TAp63 can act as a tumor suppressor in a similar manner to p53, including induction of apoptosis, cell cycle arrest, and senescence (22). ΔNp63 is selectively distributed in the basal cells of stratified and glandular epithelia and believed to be required to maintain the self-renewing capacity of stem cells in epithelial tissues, as well as drive the differentiation program of luminal cells (10, 22). The antibody used for analysis of TP63 protein levels in cells and tissue specifically targeted the transactivation domain of TP63. Therefore, the finding that lower levels of p63-transactivating isomer are associated with increased growth rate and more rapid progression through the cell cycle in shClock-transfected cells is expected.
We also analyzed the impact of decreased abundance of CLOCK on the differentiation of shClock transfected HC11 cells, including the expression of e-cadherin (CDH1), fatty acid synthase (FASN), and β-casein (CSN2). CDH1 functions as an adherens junction protein. Ductal and alveolar epithelial cells express CDH1 in the mammary, and its abundance is increased with differentiation of the gland (17). When the function of CDH1 is disrupted in adult mouse mammary gland by means of induction of a mammary-specific dominant-negative CDH1 molecule, there is massive apoptosis of alveolar cells at parturition with concomitant loss of milk production (3). Temporal analysis of CDH1 steady-state mRNA levels in the rat kidney showed it exhibits a circadian rhythm of gene expression (34), and thus, supports that CDH1 is a direct or indirect target of circadian clock regulation. In our study, we found that abundance of CDH1 mRNA and protein levels were dramatically decreased in HC11 cells transfected with shClock. Thus, disruption of clocks may negatively impact formation of adherens junctions among mammary epithelial cells and result in poorer milk production.
Reducing CLOCK levels in HC11 cells significantly decreased expression of FASN in both undifferentiated and differentiated states. FASN encodes the fatty acid synthase enzyme, which regulates milk-fat synthesis. Our previous studies found that FASN showed a circadian pattern of expression in wild-type mice during lactation (5). We also found disrupting the circadian timing system by continuously shifting light-dark cycles decreased milk-fat yield in dairy cattle (4). Together, these data support that FASN is a direct or indirect target of the BMAL1-CLOCK transcription factor in the mammary gland and that disruption of circadian clocks has a negative impact on milk fat synthesis, which may explain, in part, the poorer lactation performance of Clock-Δ19 mice.
Reducing CLOCK abundance in HC11 cells did not impact CSN2 expression. CSN2 encodes for the milk protein β-casein, and this finding suggests that the mammary clock does not directly regulate its expression. This is an interesting finding, as our studies of the impact of light-dark phase shifts on milk production in dairy cows showed decreased levels of CSN2 expression in mammary, suggesting that environmental disruption of circadian system may impact its regulation (4). Others reported that milk protein levels show circadian rhythms that are impacted by timing of feed intake (29), and thus, daily variation in substrate availability may affect milk protein synthesis. Together these studies illustrate the complexity of the circadian system from the role of local clocks, to feeding time, to systemic influences on timing of circadian rhythms and gene expression.
It is also important to discuss implications of model used in our studies that aimed to understand the role of CLOCK in the regulation of mammary epithelial growth and differentiation. Because of their importance in physiology, adaptation, and survival, most of the core circadian clock genes exist as paralogs in mice (Per1 and Per2, Cry1 and Cry2; and Clock and Npas2), and in vivo studies have shown that both genes of the pair must be knocked out to confer arhythmicity in the animals (18). Further, the interrelated/interregulated nature of the transcription-translation feedback-loop that defines the circadian clock mechanism, predisposes that the knock-down or knockout of one element affects the dynamics of others. Western blot analysis of PER2 and BMAL1 found only modest differences in levels between wild-type HC11 and shCLOCK lines. However, the dynamic changes that occurred in BMAL1 and PER2 proteins following 96 h of lactogen treatment were impacted by shCLOCK-3, with lack of a decrease in PER2 abundance after lactogen treatment. Thus, our findings on the impact of decreased CLOCK abundance on HC11 cell growth and differentiation may not be specifically due to alterations in CLOCK, but also changes in mammary circadian clock functional activity. Further, studies in a culture system eliminate systemic influences and feedback mechanisms that may be impacted or maintained in an organismal system, and, thus, future, more comprehensive studies are needed to understand the role of circadian clocks in regulation of lactation competency.
Perspectives and Significance
Our previous studies and work of others demonstrated that the mammary clock shows dynamic changes with changes in physiology from pregnancy to lactation. As the gland transitions from pregnancy to lactation, it changes from a primarily proliferative state to a differentiated state. Dynamic changes during the transition from pregnancy to lactation are marked by a relative increase in BMAL1-CLOCK abundance across the circadian cycle. Here, we demonstrate that CLOCK plays a role in regulation of mammary epithelial growth and differentiation, and report that when relative abundance of CLOCK is low, proliferation rates of mammary epithelial cells are high and levels of differentiation markers are decreased. Our study supports the hypothesis that dynamic changes in mammary core clock genes associated with physiological state change regulate developmental changes in the gland and states of proliferation and differentiation. Alterations in clock dynamics may affect lactation competency.
GRANTS
Grant support was received by the Binational Agricultural Research Development (BARD) Research Project US-4715-14, a National Institutes of Health Grant R-031R03HD-062692-01A1, a Purdue Development team Award, and an FPU 12/06079 Scholarship.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
T.C. and K.P. conception and design of research; T.C., J.C., A.S.-T., E.E., B.W., K.C., S.C., and Y.C. performed experiments; T.C., J.C., A.S.-T., E.E., B.W., K.C., S.C., A.S., S.J.M., and K.P. analyzed data; T.C., J.C., A.S.-T., E.E., B.W., S.C., A.S., S.J.M., and K.P. interpreted results of experiments; T.C. and S.C. prepared figures; T.C. drafted manuscript; T.C., J.C., A.S.-T., E.E., B.W., S.C., S.J.M., and K.P. edited and revised manuscript; T.C., J.C., A.S.-T., E.E., B.W., K.C., S.C., Y.C., A.S., S.J.M., and K.P. approved final version of manuscript.
ACKNOWLEDGMENTS
The researchers thank Dr. Marina Antoch of Roswell Park Institute for help and support with animal experiments performed.
REFERENCES
- 1.Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol 12: 540–550, 2002. doi: 10.1016/S0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 2.Antoch MP, Song E-J, Chang A-M, Vitaterna MH, Zhao Y, Wilsbacher LD, Sangoram AM, King DP, Pinto LH, Takahashi JS. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell 89: 655–667, 1997. doi: 10.1016/S0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boussadia O, Kutsch S, Hierholzer A, Delmas V, Kemler R. E-cadherin is a survival factor for the lactating mouse mammary gland. Mech Dev 115: 53–62, 2002. doi: 10.1016/S0925-4773(02)00090-4. [DOI] [PubMed] [Google Scholar]
- 4.Casey T, Crodian J, Donkin SS, Plaut K. Continuously changing light-dark phase decreases milk yield, fat, protein and lactose in dairy cows. J Adv Dairy Res 2: e110, 2014. doi: 10.4172/2329-888X.1000119. [DOI] [Google Scholar]
- 5.Casey TM, Crodian J, Erickson E, Kuropatwinski KK, Gleiberman AS, Antoch MP. Tissue-specific changes in molecular clocks during the transition from pregnancy to lactation in mice. Biol Reprod 90: 127, 2014. doi: 10.1095/biolreprod.113.116137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DePeters EJ, Hovey RC. Methods for collecting milk from mice. J Mammary Gland Biol Neoplasia 14: 397–400, 2009. doi: 10.1007/s10911-009-9158-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dolatshad H, Campbell EA, O’Hara L, Maywood ES, Hastings MH, Johnson MH. Developmental and reproductive performance in circadian mutant mice. Hum Reprod 21: 68–79, 2006. doi: 10.1093/humrep/dei313. [DOI] [PubMed] [Google Scholar]
- 8.Domínguez-Kelly R, Martín Y, Koundrioukoff S, Tanenbaum ME, Smits VA, Medema RH, Debatisse M, Freire R. Wee1 controls genomic stability during replication by regulating the Mus81-Eme1 endonuclease. J Cell Biol 194: 567–579, 2011. doi: 10.1083/jcb.201101047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol 12: 551–557, 2002. doi: 10.1016/S0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 10.Forster N, Saladi SV, van Bragt M, Sfondouris ME, Jones FE, Li Z, Ellisen LW. Basal cell signaling by p63 controls luminal progenitor function and lactation via NRG1. Dev Cell 28: 147–160, 2014. doi: 10.1016/j.devcel.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 111: 41–50, 2002. doi: 10.1016/S0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 12.Hardin PE. Transcription regulation within the circadian clock: the E-box and beyond. J Biol Rhythms 19: 348–360, 2004. doi: 10.1177/0748730404268052. [DOI] [PubMed] [Google Scholar]
- 13.Harmar AJMH, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, Kelly JS, Maywood ES, Hastings MH. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell 109: 497–508, 2002. doi: 10.1016/S0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- 14.Hoshino K, Wakatsuki Y, Iigo M, Shibata S. Circadian Clock mutation in dams disrupts nursing behavior and growth of pups. Endocrinology 147: 1916–1923, 2006. doi: 10.1210/en.2005-1343. [DOI] [PubMed] [Google Scholar]
- 15.Kennaway DJBM, Boden MJ, Voultsios A. Reproductive performance in female ClockΔ19 mutant mice. Reprod Fertil Dev 16: 801–810, 2004. doi: 10.1071/RD04023. [DOI] [PubMed] [Google Scholar]
- 16.King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TDL, Vitaterna MH, Kornhauser JM, Lowrey PL, Turek FW, Takahashi JS. Positional cloning of the mouse circadian clock gene. Cell 89: 641–653, 1997. doi: 10.1016/S0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knudsen KA, Wheelock MJ. Cadherins and the mammary gland. J Cell Biochem 95: 488–496, 2005. doi: 10.1002/jcb.20419. [DOI] [PubMed] [Google Scholar]
- 18.Kondratov RV, Gorbacheva VY, Antoch MP. The role of mammalian circadian proteins in normal physiology and genotoxic stress responses. Curr Top Dev Biol 78: 173–216, 2007. doi: 10.1016/S0070-2153(06)78005-X. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408, 2001. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Maningat PD, Sen P, Rijnkels M, Sunehag AL, Hadsell DL, Bray M, Haymond MW. Gene expression in the human mammary epithelium during lactation: the milk fat globule transcriptome. Physiol Genomics 37: 12–22, 2009. doi: 10.1152/physiolgenomics.90341.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science 302: 255–259, 2003. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 22.Melino G, Memmi EM, Pelicci PG, Bernassola F. Maintaining epithelial stemness with p63. Sci Signal 8: re9, 2015. doi: 10.1126/scisignal.aaa1033. [DOI] [PubMed] [Google Scholar]
- 23.Metz RP, Qu X, Laffin B, Earnest D, Porter WW. Circadian clock and cell cycle gene expression in mouse mammary epithelial cells and in the developing mouse mammary gland. Dev Dyn 235: 263–271, 2006. doi: 10.1002/dvdy.20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oster H, Damerow S, Kiessling S, Jakubcakova V, Abraham D, Tian J, Hoffmann MW, Eichele G. The circadian rhythm of glucocorticoids is regulated by a gating mechanism residing in the adrenal cortical clock. Cell Metab 4: 163–173, 2006. doi: 10.1016/j.cmet.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109: 307–320, 2002. doi: 10.1016/S0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 26.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109: 307–320, 2002. doi: 10.1016/S0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 27.Potten CS, Booth D, Cragg NJ, Tudor GL, O’Shea JA, Appleton D, Barthel D, Gerike TG, Meineke FA, Loeffler M, Booth C. Cell kinetic studies in the murine ventral tongue epithelium: thymidine metabolism studies and circadian rhythm determination. Cell Prolif 35, Suppl 1: 1–15, 2002. doi: 10.1046/j.1365-2184.35.s1.1.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, O’Neill JS, Wong GKY, Chesham J, Odell M, Lilley KS, Kyriacou CP, Hastings MH. Circadian orchestration of the hepatic proteome. Curr Biol 16: 1107–1115, 2006. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 29.Rottman LW, Ying Y, Zhou K, Bartell PA, Harvatine KJ. The daily rhythm of milk synthesis is dependent on the timing of feed intake in dairy cows. Physiol Rep 2: e12049, 2014. doi: 10.14814/phy2.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitt B, Povinelli L, Crodian J, Casey T, Plaut K. Circadian rhythms of ewes suckling singletons versus twins during the second week of lactation. BIOS 85: 207–217, 2014. doi: 10.1893/0005-3155-85.4.207. [DOI] [Google Scholar]
- 31.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature 417: 78–83, 2002. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 32.Ungar F, Halberg F. Circadian rhythm in the in vitro response of mouse adrenal to adrenocorticotropic hormone. Science 137: 1058–1060, 1962. doi: 10.1126/science.137.3535.1058. [DOI] [PubMed] [Google Scholar]
- 33.Roth V. Doubling time computing. http://www.doubling-time.com/compute.php. 2006.
- 34.Yamato M, Ito T, Iwatani H, Yamato M, Imai E, Rakugi H. E-cadherin and claudin-4 expression has circadian rhythm in adult rat kidney. J Nephrol 23: 102–110, 2010. [PubMed] [Google Scholar]