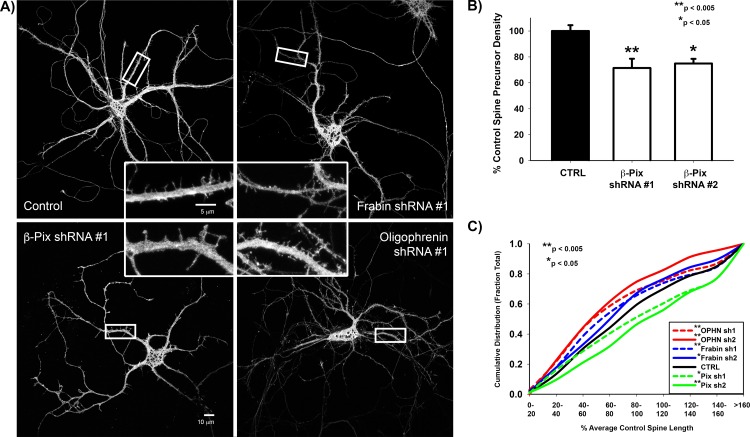
Fig 2. Regulators of spine precursor formation and elongation.
A) Representative Images of GFP-expressing DIV-9 neurons transfected with the indicated shRNA targeting sequence for 72 hours. B) β-pix shRNA targeting sequences significantly decrease spine precursor density in DIV-9/10 neurons. Spine density is expressed as the percentage of the average control spine density. n = 24 control neurons, 15 β-PIX shRNA #1 neurons, and 7 β-PIX shRNA #2 neurons; p = 0.001 for Control vs β-pix shRNA #1 (t-test) and p = 0.007 for Control vs β-pix shRNA #2 (t-test). C) Cumulative distribution plot of spine length in DIV-9/10 primary rat hippocampal neurons co-expressing GFP and the indicated shRNA targeting sequence. Spine length is expressed as a percentage of the average control spine length. shRNAs against either the RhoA-GAP, Oligophrenin-1, or Cdc42-GEF, Frabin, significantly decrease spine length, whereas shRNAs against β-pix significantly increase spine length. n = 1114 control, 680 Oligophrenin-1 shRNA #1, 115 Oligophrenin-1 shRNA #2, 613 Frabin shRNA #1, 393 Frabin shRNA #2, 465 β-pix shRNA #1, and 166 β-pix shRNA #2 spines; p = 0.005 for Control vs β-pix shRNA #1 (Mann-Whitney Rank Sum Test), p < 0.001 for Control vs β-pix shRNA #2 (Mann-Whitney Rank Sum Test), p = 0.001 for Control vs Frabin shRNA #1 (Mann-Whitney Rank Sum Test), p = 0.01 for Control vs Frabin shRNA #2 (Mann-Whitney Rank Sum Test), p < 0.001 for Control vs Oligophrenin-1 shRNA #1 (Mann-Whitney Rank Sum Test), p < 0.001 for Control vs Oligophrenin-1 shRNA #2 (Mann-Whitney Rank Sum Test).