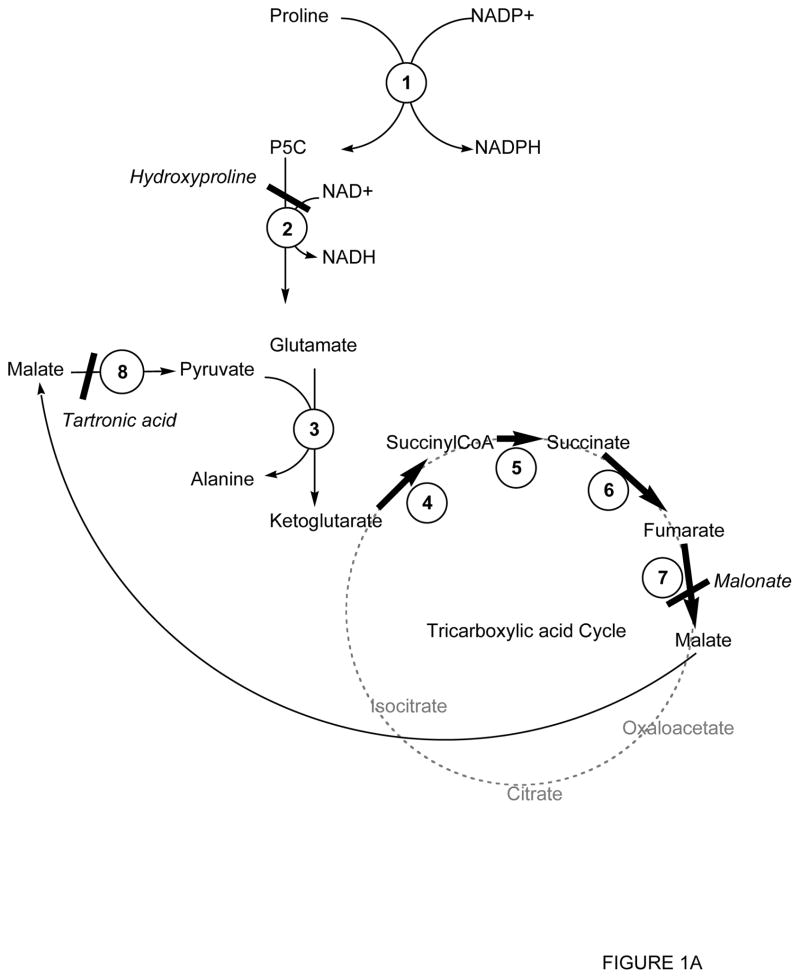
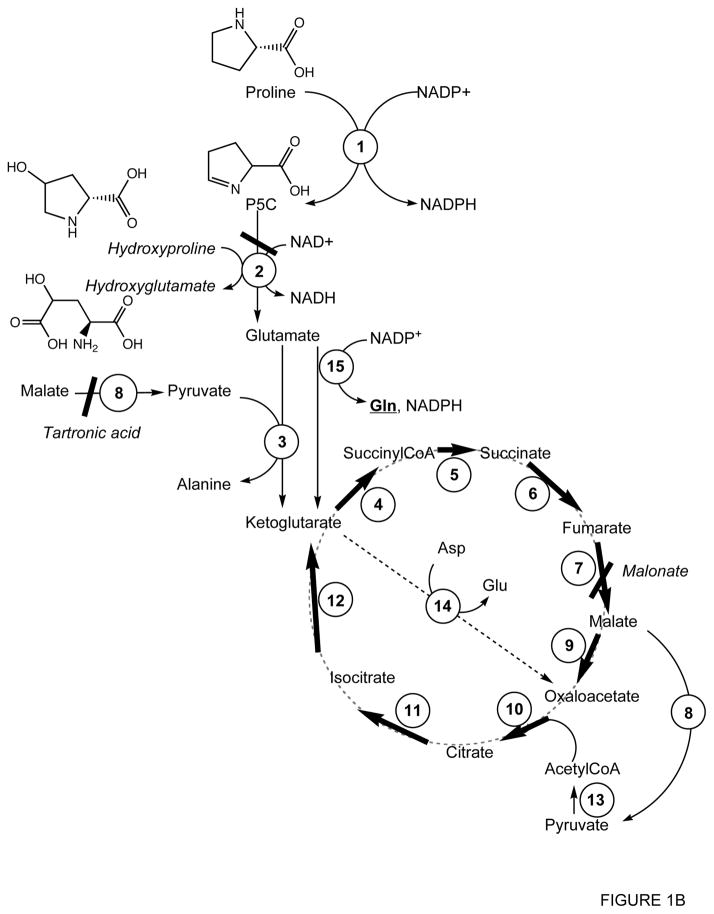
Figure 1.
Schematic representation of proline metabolism in ASE mitochondria
Panel A: Proline metabolism as described by [42–44, 47, 71].
Panel B: Proline metabolism expanded and modified according to the experimental results found in this study.
Inhibitors are shown in italics. 1, Δ1-pyrroline-5-carboxylate reductase; 2, Δ1-pyrroline-5-carboxylate dehydrogenase; 3, Glutamate-pyruvate transaminase; 4, ketoglutarate dehydrogenase; 5, succinylCoA synthetase; 6, succinate dehydrogenase; 7, fumarate reductase; 8, malic enzyme; 9, malate dehydrogenase; 10, citrate synthase; 11, aconitase; 12, isocitrate dehydrogenase; 13, pyruvate dehydrogenase; 14, aspartate-oxaloacetate transaminase; 15, glutamate dehydrogenase.