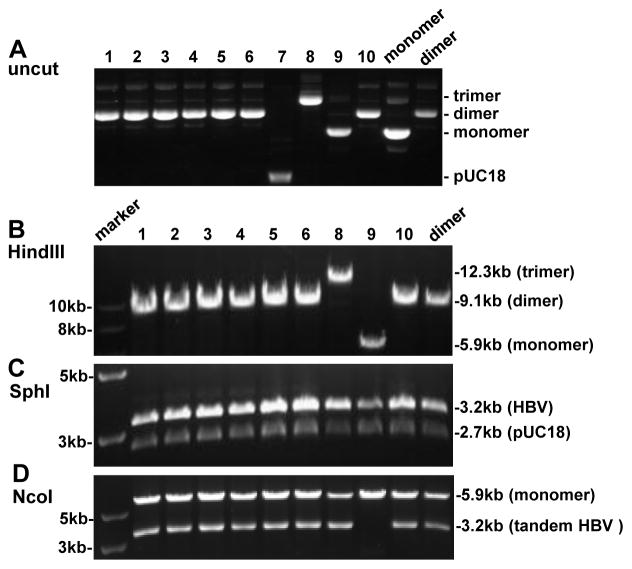
Fig. 3.
Screening for SphI dimers generated by SphI partial digestion of SphI monomer. (A) Uncut DNA from 10 clones with uncut dimer and monomer serving as controls. Clone 7 is probably pUC18 DNA. (B) HindIII digestion of 9 clones with HindIII cut dimer serving as control. The expected size is 9.1 kb for a dimer and 5.9 kb for a monomer. (C) SphI digestion, which will convert both a dimer and a monomer into 3.2-kb and 2.7-kb bands. (D) NcoI digestion. A 3.2-kb band will be released only when a dimer (or trimer) is tandem instead of tail-to-tail or head-to-head.