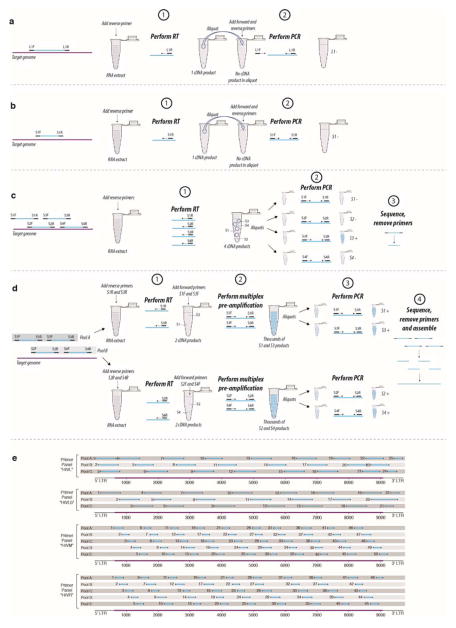
Extended Data Figure 1. Jackhammering schematic and primer panels and pools.
a through d: detection and amplification of target RNA molecules in old, degraded, low-titre samples. For the purposes of illustration, consider a tube with 1013 RNA molecules, but (because of the low RNA quality) only one molecule that is (i) capable of being primed by the given reverse primer(s) and (ii) long enough to form a 200bp product. a, conventional RT-PCR with a long amplification product, oversized for a sample with RNA less than ~200 bases in length. b, RT-PCR with a shorter amplification product. c, use of multiple primer pairs to increase the chance of a at least one PCR-positive result. d, the jackhammering approach, which overcomes the problems encountered in a through c by (i) targeting an extensive panel of short amplicons appropriately sized to the level of RNA survival in the sample, (ii) conducting RT with pools of primer pairs that amplify discrete, non-overlapping genomic regions, and (iii) employing a multiplex pre-amplification step, in the tube with the RT product, to generate sufficient DNA to ensure that each aliquot from it contains numerous template molecules for final PCR amplification. In this schematic, we show just two primer pairs per pool, but we used pools of ten pairs with our largest primer panels (shown in panel e, HXB2 numbering along HIV-1 genome). With a 10 primer-pair pool, and 10 final reactions, one can reliably recover 10 bands for sequencing. Five such pools (one entire panel of 50 pairs), allows for complete HIV-1 genome recovery even in heavily degraded samples.