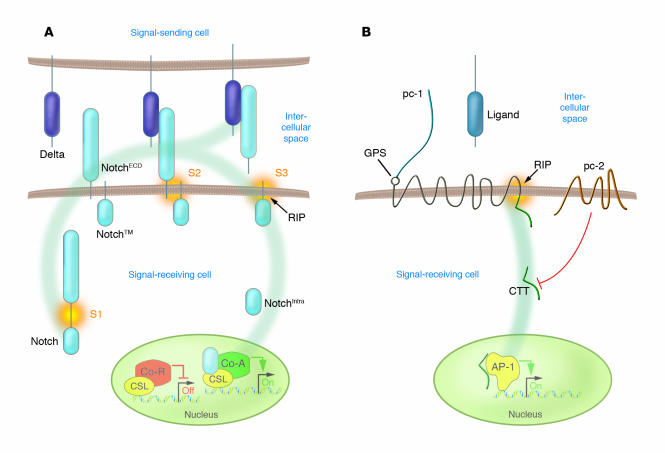
Figure 1.
Models of Notch and polycystin-1 signaling via RIP. (A) Newly synthesized Notch is constitutively cleaved in the trans-Golgi network by furin-like proteases. Following this site 1 (S1) cleavage event, the bipartite Notch receptor consisting of a noncovalent interaction between the ectodomain (NotchECD) and a membrane-tethered intracellular domain (NotchTM) is inserted into the plasma membrane. Activation of Notch by its ligand, e.g., Delta, triggers 2 additional proteolytic events. S2 cleavage by extracellular proteases of the ADAM/TACE (a disintegrin and metalloproteinase/TNF-––converting enzyme) family releases NotchECD and generates an activated, membrane-bound form of NotchTM that is further processed via RIP-mediated S3 cleavage. These events lead to release of the Notch intracellular domain (NotchIntra), which translocates into the nucleus and displaces the corepressor (Co-R) complex from the CBF-1, Su(H), Lag-1–type (CSL-type) transcription factors. The coactivating (Co-A) complex containing NotchIntra and CSL stimulates expression of CSL/Notch target genes. Figure modified from Development (17) with permission from the Company of Biologists Ltd.; and from Current Biology (18) with permission from Elsevier. (B) Polycystin-1 (pc-1) is a cell-surface receptor that undergoes regulated extracellular proteolytic processing at its GPS, which results in the release of its N-terminal fragment. In this issue of the JCI, Chauvet et al. (14) show that the CTT of polycystin-1 can also be cleaved from its transmembrane anchor, presumably through RIP-related mechanisms. Once released, this domain translocates to the nucleus, where it activates the AP-1 transcription pathway. This translocation event appears to be regulated by polycystin-2.