Abstract
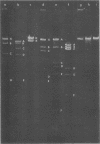
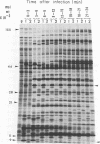
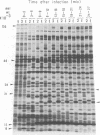
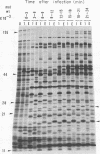
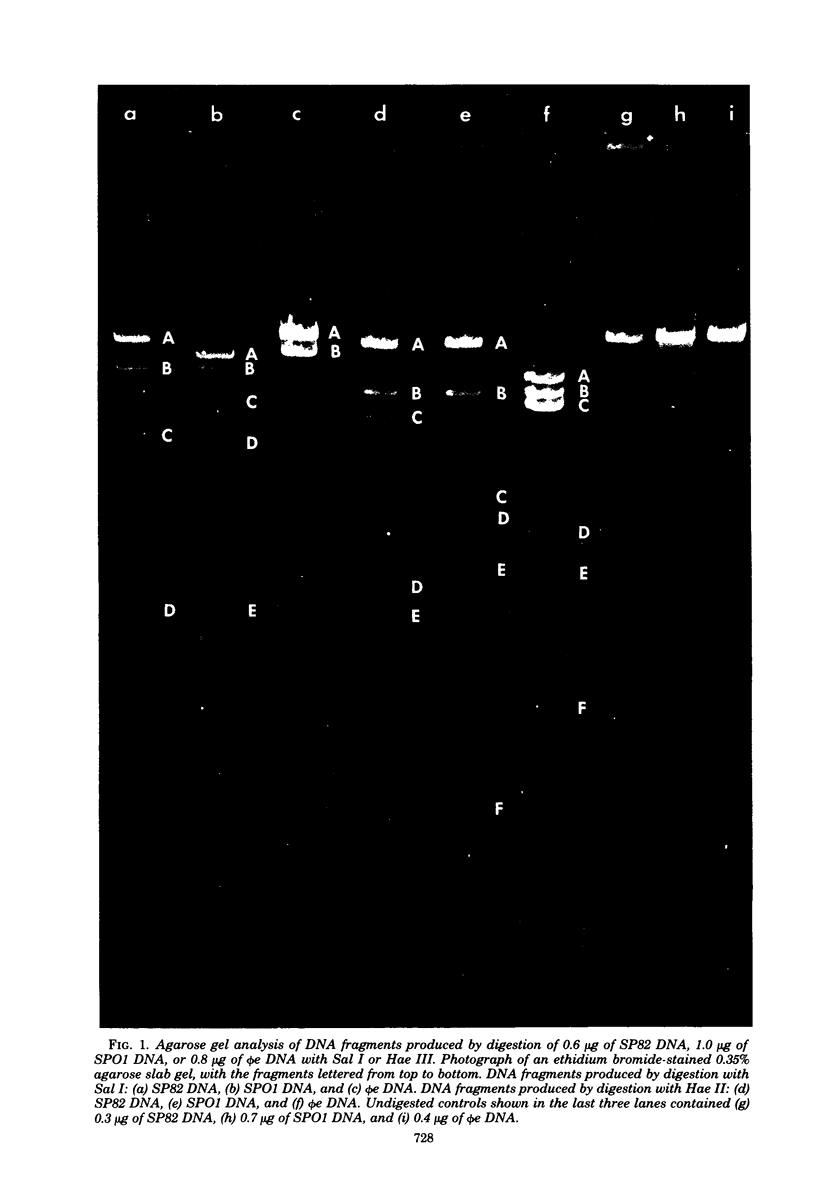
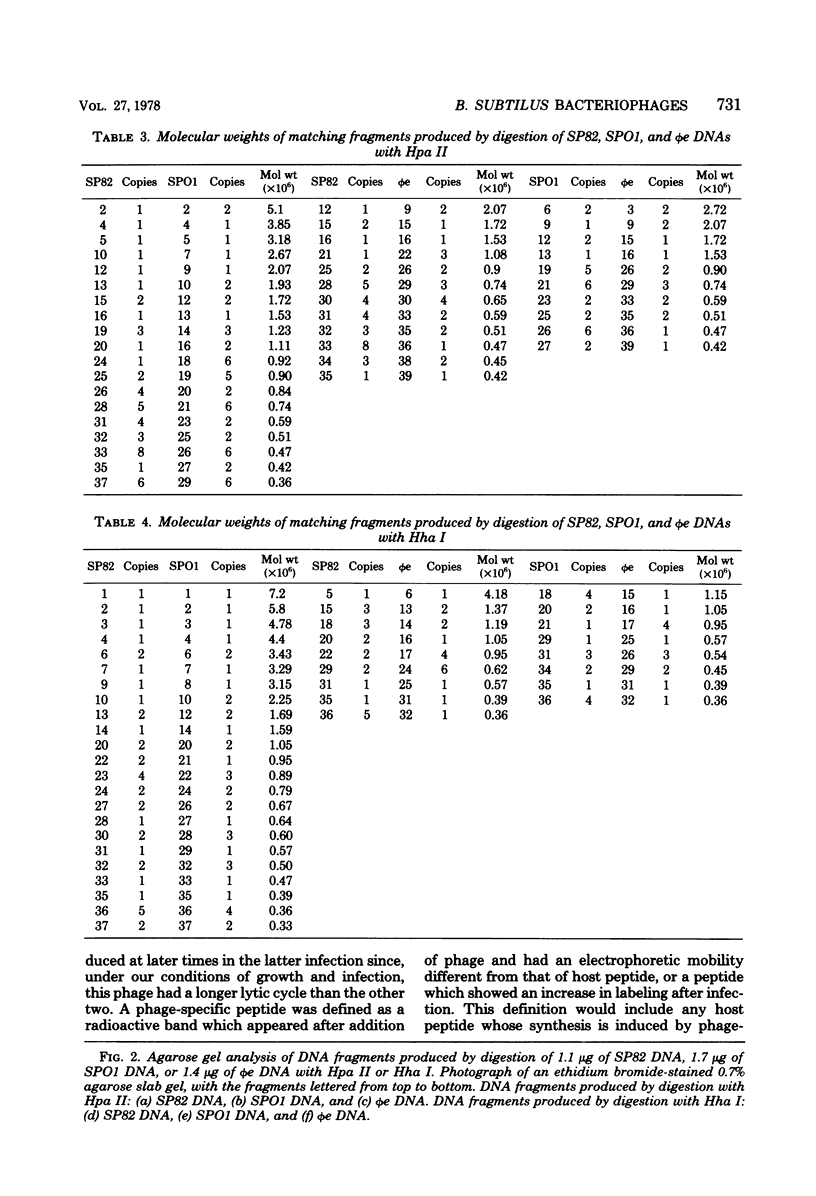
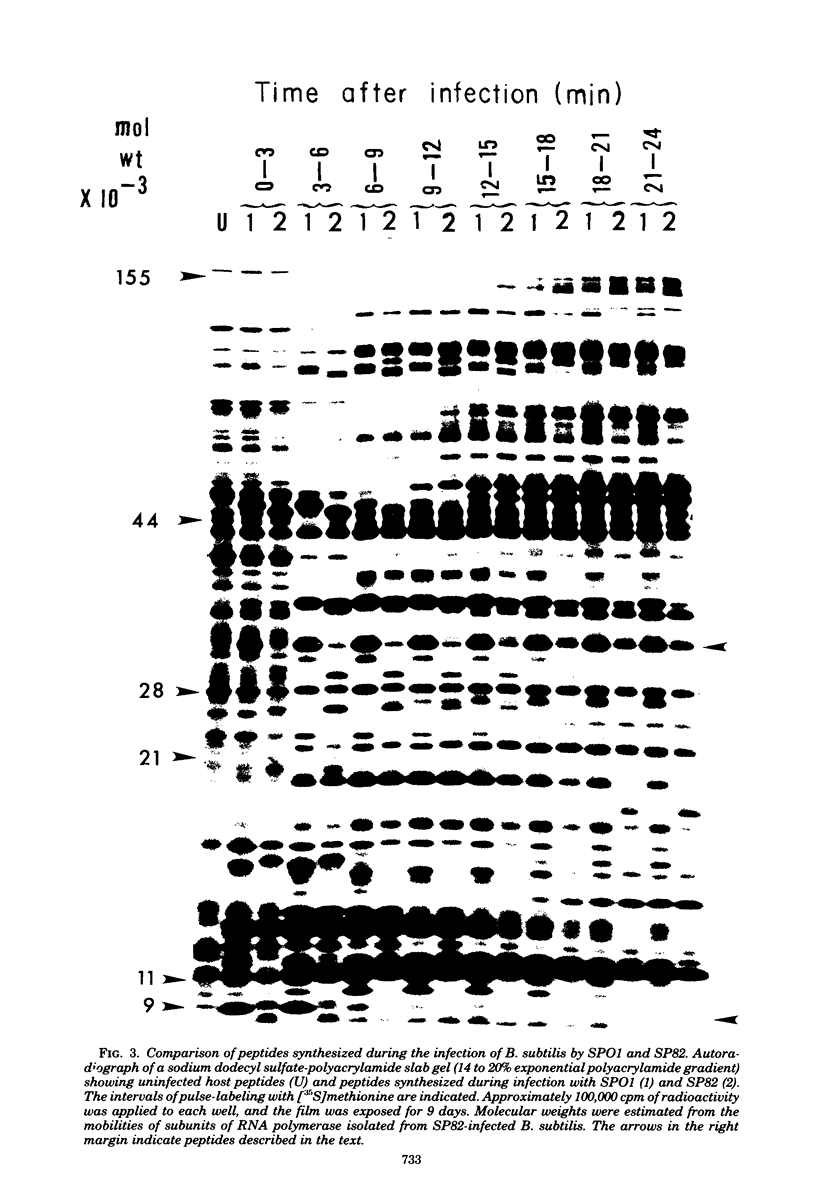
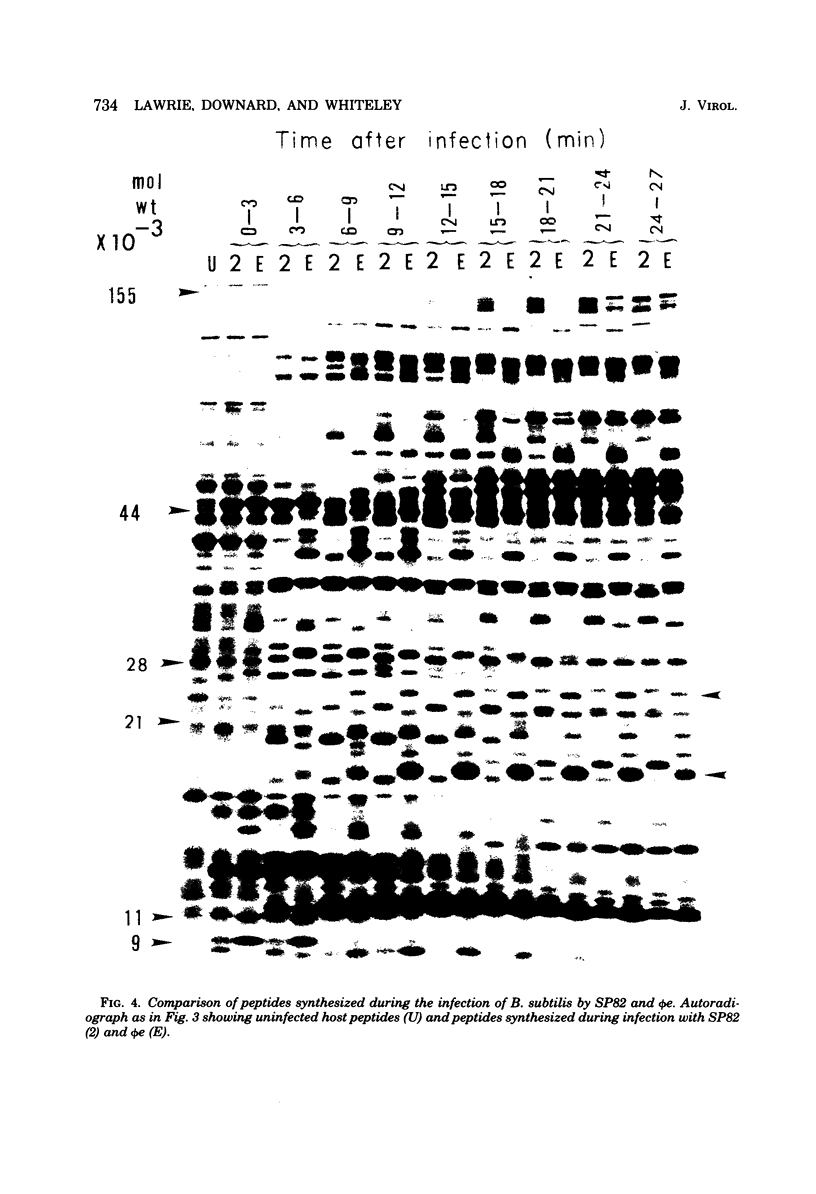
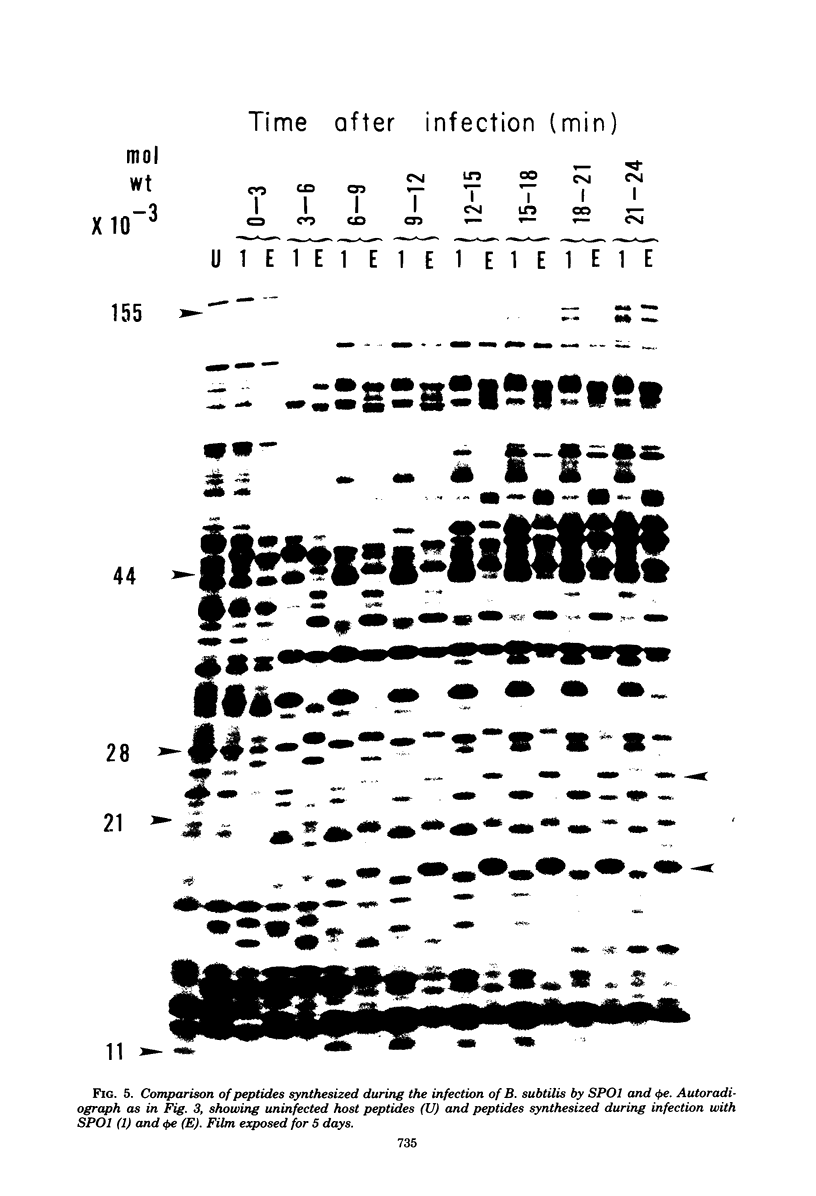
The genomes of Bacillus subtilis phages phie, SPO1, and SP82 were compared by DNA-DNA hybridization, analysis of DNA fragments produced by digestion with restriction endonucleases, comparison of the arrays of peptides synthesized during infection, and phage neutralization. DNA-DNA hybridization experiments indicated that about 78% of the SP82 DNA was homologous with SPO1 DNA, whereas 40% of the phie DNA was homologous to either SPO1 or SP82 DNA. Agarose gel electrophoresis was used to compare the molecular weights of DNA fragments produced by cleavage of SP82, SPO1, and phie DNAs with the restriction endonucleases Hae III, Sal I, Hpa II, and Hha I. Digestion of the DNAs with Hae III and Sal I produced only a few fragments, whereas digestion with Hpa II and Hha I yielded 29 to 40 fragments, depending on the DNA and the enzyme. Comparing the Hpa II fragments, 51% of the SP82 fragments had mobilities which matched those of SPO1 fragments, 32% of the SP82 fragments matched the phie fragments, and 34% of the SPO1 fragments matched the phie fragments. Comparing the Hha I digestion products, 62% of the SP82 fragments had mobilities matching the SPO1 fragments, 24% of the SP82 fragments matched the phie fragments, and 22% of the SPO1 fragments matched the phie fragments. Analysis of peptides by electrophoresis on one-dimensional sodium dodecyl sulfate-polyacrylamide slab gels showed that approximately 70 phage-specific peptides were synthesized in the first 24 min of each infection. With mobility and the intervals of synthesis as criteria, 66% of the different SP82 peptides matched the SPO1 peptides, 34% of the SP82 peptides matched the phie peptides, and 37% of the SPO1 peptides matched the phie peptides. Phage neutralization assays using antiserum to SP82 yielded K values of 510 for SP82, 240 for SPO1, and 120 for phie.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brodetsky A. M., Romig W. R. Characterization of Bacillus subtilis bacteriophages. J Bacteriol. 1965 Dec;90(6):1655–1663. doi: 10.1128/jb.90.6.1655-1663.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosa J. H., Brenner D. J., Falkow S. Use of a single-strand specific nuclease for analysis of bacterial and plasmid deoxyribonucleic acid homo- and heteroduplexes. J Bacteriol. 1973 Sep;115(3):904–911. doi: 10.1128/jb.115.3.904-911.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Duffy J. J., Geiduschek E. P. Purification of a positive regulatory subunit from phage SP01-modified RNA polymerase. Nature. 1977 Nov 3;270(5632):28–32. doi: 10.1038/270028a0. [DOI] [PubMed] [Google Scholar]
- Duffy J. J., Petrusek R. L., Geiduschek E. P. Conversion of Bacillus subtilis RNA polymerase activity in vitro by a protein induced by phage SP01. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2366–2370. doi: 10.1073/pnas.72.6.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox T. D. Identification of phage SP01 proteins coded by regulatory genes 33 and 34. Nature. 1976 Aug 26;262(5571):748–753. doi: 10.1038/262748a0. [DOI] [PubMed] [Google Scholar]
- Fox T. D., Pero J. New phage-SPO1-induced polypeptides associated with Bacillus subtilis RNA polymerase. Proc Natl Acad Sci U S A. 1974 Jul;71(7):2761–2765. doi: 10.1073/pnas.71.7.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage L. P., Geiduschek E. P. RNA synthesis during bacteriophage SPO1 development: six classes of SPO1 RNA. J Mol Biol. 1971 Apr 28;57(2):279–297. doi: 10.1016/0022-2836(71)90346-9. [DOI] [PubMed] [Google Scholar]
- Hemphill H. E., Whiteley H. R. Bacteriophages of Bacillus subtilis. Bacteriol Rev. 1975 Sep;39(3):257–315. doi: 10.1128/br.39.3.257-315.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiatt W. R., Whiteley H. R. Translation of RNAs synthesized in vivo and in vitro from bacteriophage SP82 DNA. J Virol. 1978 Feb;25(2):616–629. doi: 10.1128/jvi.25.2.616-629.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie J. M. DNA strand specificity of transcripts produced in vivo and in vitro by RNA polymerase from SP82-infected Bacillus subtilis. J Virol. 1975 May;15(5):1286–1288. doi: 10.1128/jvi.15.5.1286-1288.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Palefski S., Hemphill H. E., Kolenbrander P. E., Whiteley H. R. Dominance relationships in mixedly infected Bacillus subtilis. J Virol. 1972 Apr;9(4):594–601. doi: 10.1128/jvi.9.4.594-601.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiewe M. H., Crosa J. H., Ordal E. J. Deoxyribonucleic acid relationships among marine vibrios pathogenic to fish. Can J Microbiol. 1977 Aug;23(8):954–958. doi: 10.1139/m77-142. [DOI] [PubMed] [Google Scholar]
- Shenk T. E., Rhodes C., Rigby P. W., Berg P. Biochemical method for mapping mutational alterations in DNA with S1 nuclease: the location of deletions and temperature-sensitive mutations in simian virus 40. Proc Natl Acad Sci U S A. 1975 Mar;72(3):989–993. doi: 10.1073/pnas.72.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman G. B., Whiteley H. R. In vivo and in vitro transcription by ribonucleic acid polymerase from SP82-infected Bacillus subtilis. J Biol Chem. 1974 Mar 10;249(5):1483–1489. [PubMed] [Google Scholar]
- Spiegelman G. B., Whiteley H. R. Purification of ribonucleic acid polymerase from SP82-infected Bacillus subtilis. J Biol Chem. 1974 Mar 10;249(5):1476–1482. [PubMed] [Google Scholar]
- Talkington C., Pero J. Restriction fragment analysis of the temporal program of bacteriophage SPO1 transcription and its control by phage-modified RNA polymerases. Virology. 1977 Dec;83(2):365–379. doi: 10.1016/0042-6822(77)90181-7. [DOI] [PubMed] [Google Scholar]
- Truffaut N., Revet B., Soulie M. O. Etude comparative des DNA de phages 2C, SP8*, SP82, phi e, SP01 et SP50. Eur J Biochem. 1970 Aug;15(2):391–400. doi: 10.1111/j.1432-1033.1970.tb01020.x. [DOI] [PubMed] [Google Scholar]
- Yasbin R. E., Maino V. C., Young F. E. Bacteriophage resistance in Bacillus subtilis 168, W23, and interstrain transformants. J Bacteriol. 1976 Mar;125(3):1120–1126. doi: 10.1128/jb.125.3.1120-1126.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]