Figure 1.
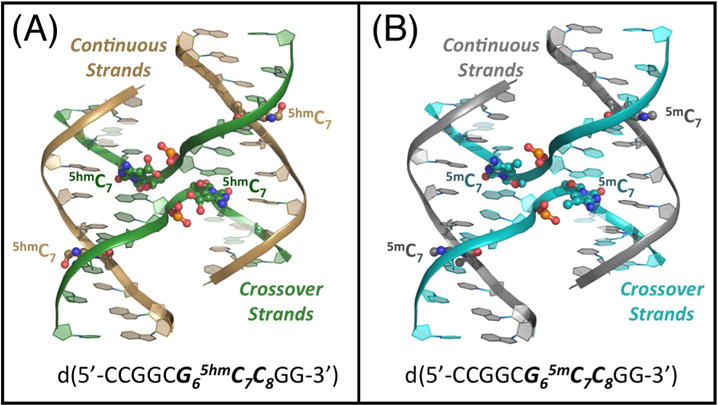
Comparison of the structures of 5-hydroxymethylcytosine (5hmC) and 5-methyl cytosine (5mC) in DNA Holliday junctions. (A) The crystal structure of the hydroxylmethylated sequence d(CCGGCG65hmC7C8GG) is shown with the DNA backbones traced as ribbons (colored gold for the outside continuous strands and green for the junction crossing strands). The 5hmC bases, along with the phosphate groups that they are H-bonded to, are rendered as ball-and-stick models, with the carbon atoms of the nucleotides along the continuous strand colored gold and those at the junction colored green. The 5hmC bases on the crossover strands have hydroxyl groups that occupy two rotamer conformations, both shown on the image. (B) The DNA backbone of the methylated sequence d(CCGGCG65mC7C8GG) is traced as ribbons (grey along the outside continuous strands and blue along the junction crossing strands). The ball-and-stick models of the 5mC bases and their interacting the phosphate groups are colored grey on the continuous and blue on the junction crossing strands.
