SUMMARY
Periodontitis is a highly prevalent disease caused in part by an aberrant host response to the oral multi-species biofilm. A balance between the oral bacteria and host immunity is essential for oral health. Imbalances in the oral microbiome lead to an uncontrolled host inflammatory response and subsequent periodontal disease (i.e. gingivitis and periodontitis). TREM-1 is a signaling receptor present on myeloid cells capable of acting synergistically with other pattern recognition receptors leading to amplification of inflammatory responses. The aim of this study was to investigate the activation of the TREM-1 pathway in the human monocyte-like cell line THP-1 exposed to both oral pathogens and commensals. The relative expression of the genes encoding TREM-1 and its adapter protein DAP12 were determined by quantitative real-time polymerase chain reaction. The surface expression of TREM-1 was determined by flow cytometry. Soluble TREM-1 and cytokines were measured by enzyme-linked immunosorbent assay. The results demonstrate that both commensal and pathogenic oral bacteria activate the TREM-1 pathway, resulting in a proinflammatory TREM-1 activity-dependent increase in proinflammatory cytokine production.
Keywords: cell receptors, innate immunity, mucosal immune responses, periodontal disease, Porphyromonas, Streptococcus
INTRODUCTION
Periodontitis is a common oral inflammatory disease leading to destruction of tooth supporting structures and alveolar bone, and, when left untreated, ultimately causes tooth loss. Periodontitis affects a substantial proportion of the population, with the prevalence of severe periodontitis estimated to be 8.5% (Eke et al., 2012). Bacteria play a critical role in initiating gingivitis and periodontitis. Several species of oral bacteria, including Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Treponema denticola and Tannerella forsythia have been associated with aggressive and chronic forms of periodontitis (Haubek et al., 2008; Oliveira et al., 2016). These bacteria stimulate host innate immune responses that play a crucial role in the pathogenesis of periodontal infection (Yucel-Lindberg & Bage, 2013). Uncontrolled host inflammatory responses can contribute to the progression of periodontitis, including effects such as severe tissue damage and loss of alveolar bone.
Triggering Receptor Expressed on Myeloid cells (TREM)-1, a member of the immunoglobulin superfamily, is a receptor expressed on monocytes and neutrophils that plays an important signaling role in the innate immune response against pathogenic bacteria, viruses and fungi (Sharif & Knapp, 2008; Buckland et al., 2011; Ivan et al., 2012). Activation of TREM-1 leads to a cascade of intracellular signaling events resulting in activation and degranulation of neutrophils, enhanced phagocytosis and cytokine production (Arts et al., 2013). TREM-1 consists of an extracellular domain, a transmembrane region, as well as a short cytoplasmic domain that lacks signaling motifs (Ramanathan et al., 2005). Consequently, the association of TREM-1 with the 12-kDa adaptor protein DNAX activation protein (DAP12) is required for induction of downstream signaling (Bouchon et al., 2000). Although TREM-1 ligands are yet to be definitively identified, putative ligands include high-mobility group box 1 (HMGB1), 70-kDa heat-shock protein (hsp 70), a ligand expressed on platelets, peptidoglycan recognition protein 1 (PGLYRP1) of neutrophils, as well as bacterial peptidoglycans and lipopolysaccharides (Bouchon et al., 2001; El Mezayen et al., 2007; Haselmayer et al., 2007; Read et al., 2015).
TREM-1 plays a major role in bacterial infection and sepsis by acting as an amplifier of inflammation and is necessary for a successful antibacterial response. In a polymicrobial sepsis mouse model, a moderate amount of TREM-1 silencing by small interfering RNA (siRNA) enhanced mouse survival, likely by reducing the systemic inflammatory response, whereas high doses of siRNA resulted in impaired neutrophil function and increased mouse mortality (Gibot et al., 2007) suggesting that optimized levels of TREM-1 are required for mounting an effective immune response and clearance pathogens. TREM-1 can activate the nuclear factor-κβ (NF-κB) pathway leading to a proinflammatory response characterized by increased production of cytokines tumor necrosis factor-α (TNF-α), interleukin-2 (IL-2), IL-12p40 and IL-1β (Gibot et al., 2004; Sharif et al., 2007). TREM-1 also modulates the signaling pathways of pattern recognition receptors, including Toll-like receptors (TLRs) and Nod-like receptors (Arts et al., 2013). For example, TREM-1 can act synergistically with TLR-2 in inducing cytokine production (Radsak et al., 2004), whereas TLR-2 concomitantly upregulates TREM-1 via the MyD88-dependent pathway (Buckland et al., 2011). Molecular cross-talk between TREM-1 and TLR-2 may also occur at the level of NF-κB transcription factor as it is involved in both pathways (Arts et al., 2013). TREM-1 additionally modulates the TLR-4 pathway (Arts et al., 2011), by regulating the expression of MyD88 and CD14, through regulation of NF-κB (Ornatowska et al., 2007), and by downregulating negative regulators of the TLR pathway (e.g. IRAK-M) (Lagler et al., 2009).
A soluble form of TREM-1 (sTREM-1) has been detected in the plasma of endotoxin-treated mice, bronchoalveolar lavage fluids from patients with pneumonia, serum of patients with septic shock and gingival crevicular fluid of patients with periodontitis (Gibot et al., 2004; Bisson et al., 2012; Palazzo et al., 2012), raising the possibility that sTREM-1 may be useful as a diagnostic marker for sepsis and inflammation. The mechanism of formation of sTREM-1 is not yet understood, although alternate splicing of TREM-1 mRNA (Gingras et al., 2002; Baruah et al., 2015) and proteolytic cleavage of membrane-bound TREM-1 by matrix metalloproteinases (Gomez-Pina et al., 2007) have been proposed as possibilities. The function of sTREM is also not clear, but when released it may competitively bind TREM-1 ligands, thereby acting as a negative regulator of the TREM-1 pathway; sTREM-1 has been shown to have a protective effect on murine septic peritonitis (Bouchon et al., 2001).
Fine-tuning of the immune response is critical in preventing excessive inflammation and tissue damage. TREM-1 receptors can act as amplifiers of the inflammatory response and cytokine production by monocytes and neutrophils. Therefore, modulating TREM-1 expression may help to dampen immune responses to benefit the host. Little is known regarding the regulation of TREM-1 expression in response to periodontal pathogens, and it is not clear how different periodontal bacteria, including Gram-negative pathogens, as well as Gram-positive commensals, activate the TREM-1 pathway in the context of periodontitis. The objective of this study was to compare the induction of TREM-1 expression and proinflammatory cytokine production in THP-1 monocytes following exposure to several species of periodontal bacteria.
MATERIALS AND METHODS
Host cells
The human monocyte-like cell line THP-1 was obtained from Sigma Aldrich (St Louis, MO). The cells were maintained in RPMI Glutamax medium (Life Technologies, Grand island, NY) supplemented with 10% fetal bovine serum (FBS; Life Technologies) at a density of 1 × 106 to 2 × 106 cells ml−1. The cells were split every 3 to 4 days. Cells were maintained in a 5% CO2 incubator at 37°C. For experiments, 1 × 106 cells per well were seeded in 12-well tissue-culture coated plates (Falcon).
Bacteria
The bacterial species A. actinomycetemcomitans, P. gingivalis, T. forsythia, Streptococcus gordonii, Streptococcus cristatus and Streptococcus pneumoniae were tested. Both the Gram-positive respiratory pathogen S. pneumoniae and the Gram-negative periodontal pathogen P. gingivalis are known positive activators of the TREM-1 pathway and were used as positive experimental controls. S. gordonii and S. cristatus were chosen to represent oral commensal members of the mitis group of streptococci that also includes S. pneumoniae. The bacterial strains obtained from our culture collection and the growth conditions used are listed in Table 1. THP-1 cells were routinely infected with bacteria at multiplicities of infection (MOI) of 10 and 100 for 4 h and 24 h. Uninfected cells were used as controls.
Table 1.
Bacterial strains and growth conditions
| Bacteria | Medium | Condition |
|---|---|---|
|
Aggregatibacter actinomycetemcomitans 283S (smooth phenotype, serotype A) |
TSBY (0.6% yeast extract and 0.04% sodium bicarbonate) |
Candle jar |
|
Porphyromonas gingivalis 381 |
Sheep blood agar, TSBY with hemin and vitamin K |
Anaerobic chamber |
|
Streptococcus gordonii G9B |
TSBY (0.5% yeast extract) |
Candle jar |
|
Streptococcus cristatus CR3 |
Todd–Hewitt agar/broth | Candle jar |
|
Streptococcus pneumoniae TIGR4 |
Blood agar, Todd-Hewitt broth |
Aerobic |
|
Tannerella forsythia 43037 |
TF agar/broth (with N-acetyl muramic acid) |
Anaerobic chamber |
TSBY, tryptic soy broth supplemented with yeast extract as indicated. TF, Tannerella forsythia.
Survival of bacteria in co-culture with THP-1 cells
To determine the survival of each bacterial strain co-cultured with the cell line, 1 × 108 bacteria were added to 1 × 106 THP-1 cells in 1 ml of cell culture medium and incubated in 5% CO2 at 37°C for 4 and 24 h. At the end of each time period, the bacterial suspension was serially diluted and spotted onto duplicate agar plates and incubated under conditions appropriate for the bacterial strain. The number of bacteria recovered was enumerated by counting colony-forming units (CFU) using the spotting–spreading method as described elsewhere (Thomas et al., 2012). Uninfected THP-1 cells were included as a control for sterility.
Cytotoxicity of bacteria on THP-1 cells
Cytotoxicity was determined using the Pierce LDH cytotoxicity assay kit (Thermo Scientific, Waltham, MA) according to the manufacturer’s instructions. Briefly, THP-1 cells were seeded in RPMI medium with 10% FBS at a density of 1 × 104 cells per well in 96-well tissue culture-coated plates (Falcon). Cells were co-incubated with bacteria at MOIs of 10 and 100, in triplicate, and incubated in 5% CO2 at 37°C for 4 and 24 h. At the end of the incubation period, the 96-well plate was centrifuged at 250 g for 3 min. The cell culture medium (50 µl) was transferred to appropriate wells of a new 96-well flat-bottomed plate (Falcon). The reaction mixture (50 µl) was added to each well and mixed by gentle tapping. The plate was incubated at room temperature for 30 min protected from light. Stop solution (50 µl well−1) was added to each sample well and mixed by gentle tapping. Absorbance was measured at 490 nm and 680 nm using the Beckman AD340 microplate reader (Beckman Coulter, Brea, CA). To determine LDH activity, absorbance value at 680 nm (background signal from the instrument) was subtracted from the 490 nm absorbance. Cytotoxicity was expressed as a percentage of the maximum LDH activity.
RNA extraction
RNA extraction was performed using the RNeasy mini kit (Qiagen, Hilden, Germany). After 4 and 24 h of infection, THP-1 cells were centrifuged, and the supernatant was removed and stored at −80°C for cytokine analysis. RNA was extracted from the cell pellet following the manufacturer’s instructions. RNA concentration and purity were determined using a Nanodrop 2000 spectrophotometer. RNA was used immediately for cDNA synthesis or stored at −80°C until further use.
Reverse transcription (cDNA synthesis)
Reverse transcription was performed using the High Capacity RNA-to-cDNA kit (Applied Biosystems, Foster City, CA) according to manufacturer’s instructions. An equal amount of RNA (1 µg) was used for all the samples. Reverse transcription was performed in a BioRad MyCycler thermocycler (BioRad, Hercules, CA) at 37°C for 60 min followed by 95°C for 5 min. Samples without reverse transcriptase (no RT) were used as negative controls. The cDNA was stored at −20°C until further use.
Quantitative real-time polymerase chain reaction
TaqMan Gene Expression Assays and the TaqMan Gene Expression Master Mix (Life Technologies) were used according to the manufacturer’s instructions. β-Actin was used as an endogenous control (TaqMan Assay ID: Hs 01060665-g1). The relative expression of TREM-1 (TaqMan Assay ID: Hs 00218624-m1) and DAP12 (TaqMan assay ID: Hs00182426-m1) genes was determined. The cDNA from uninfected cells was used as a control. Polymerase chain reaction (PCR) conditions used were 50°C for 2 min, 95°C for 5 min followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. Quantitative PCR was carried out in a 7500 Real-time PCR System (Applied Biosystems). Data were analyzed by the ΔΔCT method using the ABI 7500 system software.
Flow cytometry
Surface expression of TREM-1 was determined by flow cytometric analysis. THP-1 cells were co-incubated with the bacteria at an MOI of 10 and 100 for 4 and 24 h. After incubation, the cells were washed with phosphate-buffered saline. Fc receptors were blocked using human FcR blocking reagent (Miltenyi Biotec, San Diego, CA). Cells were stained using mouse anti-human TREM-1 antibody (Abcam, Cambridge, MA) followed by goat anti-mouse IgG Alexa Fluor 488 secondary antibody (Abcam). Mouse IgG1 monoclonal antibody (Abcam) was used as an isotype control. After washing, the cells were resuspended in phosphate-buffered saline with 0.5% bovine serum albumin. Cells were fixed in 2% formaldehyde in FACS buffer and analyzed using a BD Fortessa flow cytometer (Becton Dickinson). The results were analyzed using FlowJo (version 8.8.7).
Cytokine analysis and measurement of sTREM-1
Levels of sTREM-1, IL-1β and TNF-α in culture supernatants were measured by enzyme-linked immunosorbent assay (ELISA) using commercially available kits (R&D Systems, Minneapolis, MN).
Effect of TREM-1 inhibitor on cytokine release
Involvement of the TREM-1 pathway in the secretion of proinflammatory cytokines was tested using the synthetic LP17 peptide (LQVTDSGLYRCVIYHPP) (Pepscan Presto B.V, Lelystad, the Netherlands) that mimics the highly conserved extracellular domain of TREM-1 believed to be involved in the binding of TREM-1 to its ligand (Radaev et al., 2003; Gibot et al., 2004). A second peptide with the same amino acids but in a different order was used as a control (LP 17 control peptide; TDSRCVIGLYHPPLQVY) (Pepscan Presto B.V). THP-1 cells were infected with the bacteria at an MOI of 100. Some wells were also treated with either LP17 or LP17 control peptide at a concentration of 100 ng ml−1. After incubating the cells for 24 h, IL-1β and TNF-α levels in the supernatant were quantified using ELISA.
Statistical analysis
Statistical analysis was performed by one-way analysis of variance followed by Tukey’s honest significant difference post hoc test. A P-value < 0.05 was considered statistically significant.
RESULTS
Cytotoxicity
A general measure of bacterial toxicity against monocytes was initially performed, with cytotoxicity determined by measuring LDH released from the THP-1 cells treated with different oral bacterial species (Table 2). Streptococcus pneumoniae induced the highest cytotoxicity and A. actinomycetemcomitans induced the least cytotoxicity.
Table 2.
Cytotoxicity induced by bacteria
| Bacteria | MOI 10 | MOI 100 |
|---|---|---|
|
Aggrega tibacter actinomycetemcomitans 283S |
1.8 ± 0.2 | 4.2 ± 0.5 |
| Porphyromonas gingivalis 381 | 3.2 ± 1.0 | 7.1 ± 0.2 |
| Streptococcus gordonii G9B | 17.0 ± 2.8 | 21.0 ± 1.9 |
| Streptococcus cristatus CR3 | 14.8 ± 1.6 | 19.0 ± 2.4 |
| Streptococcus pneumoniae TIGR4 | 26.0 ± 4.8 | 59.0 ± 2.9 |
| Tannerella forsythia 43037 | 7.9 ± 0.9 | 11.5 ± 0.4 |
THP-1 cells were exposed to indicated bacteria at multiplicities of infection (MOIs) of 10 and 100 for 24 h. Cytotoxicity was detected by measuring the LDH release from the cells and is expressed as percentage of the total LDH activity upon lysis of uninfected cells. Data shown are mean ± SD from triplicate cultures of a representative experiment.
Bacterial survival in cell culture conditions
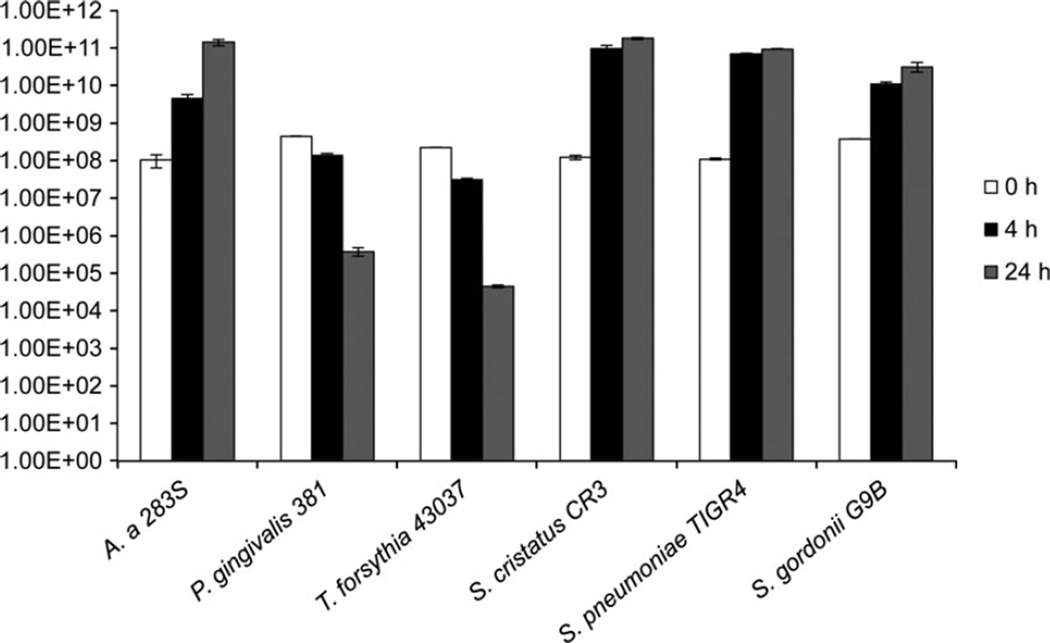
We next examined the ability of the bacteria to survive in co-culture with THP-1 cells in RPMI medium with 10% FBS in 5% CO2 at 37°C. This test was performed to make sure that the strict anaerobes (P. gingivalis and T. forsythia) would be able to survive under cell culture conditions. Aerobic and microaerophilic bacteria (S. gordonii, S. cristatus, S. pneumoniae and A. actinomycetemcomitans) survived well under cell culture conditions and increased in numbers during the 24 h period. However, as was expected, the strict anaerobes (P. gingivalis and T. forsythia) showed significant decreases in CFUs recovered after 4 and 24 h of incubation under THP-1 cell culture conditions (Fig. 1). There was no difference between the CFUs recovered when the bacteria were cultured alone (data not shown) or in co-culture with the THP-1 cells for all the bacteria tested.
Figure 1.
Survival of bacteria under co-incubation with THP-1 cell culture conditions. THP-1 cells were exposed to bacteria at multiplicity of infection (MOI) of 100. After 4 and 24 h post exposure the CFUs were determined. Experiments were repeated three times. Data shown are mean ± SD of triplicate cultures from a representative experiment. A. a., Aggregatibacter actinomycetemcomitans.
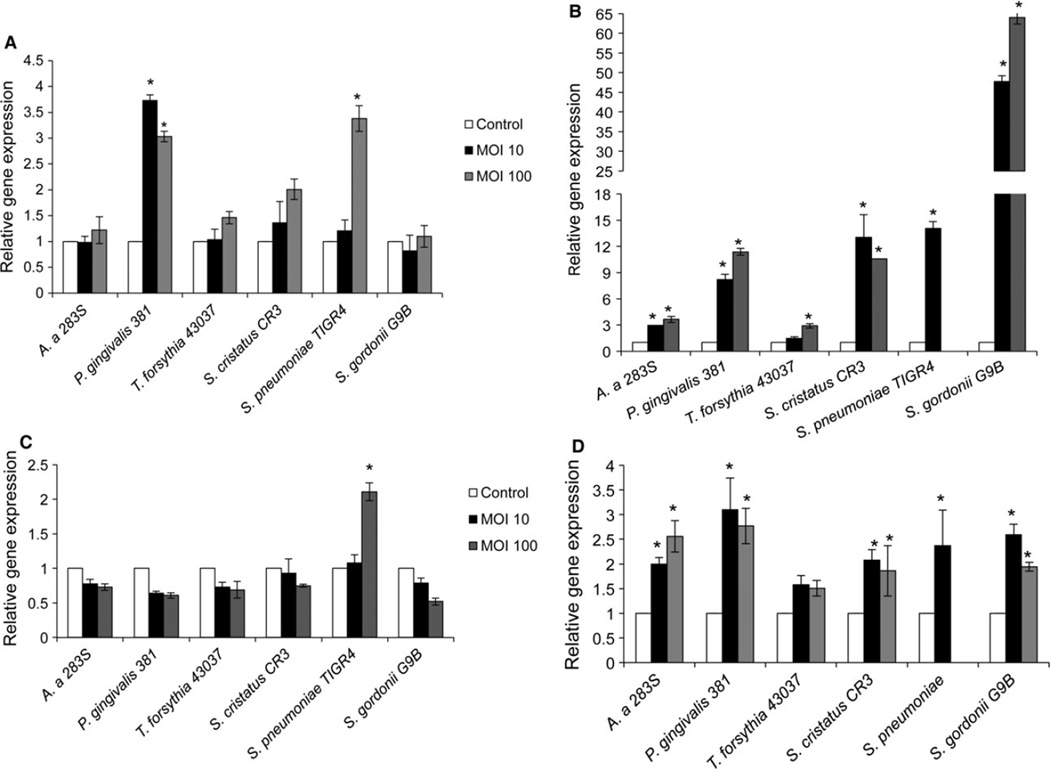
TREM-1 and DAP12 gene expression
Patterns of TREM-1 and DAP12 expression were next determined by qPCR in THP-1 cells co-incubated with different oral bacteria at an MOI of 10 and 100 for 4 or 24 h (Fig. 2). At 4 h exposure, P. gingivalis (MOI 10 and 100) and S. pneumoniae (MOI 100), two well-known pathogens, induced significantly higher expression of TREM-1 mRNA (Fig. 2A). Following 24 h of exposure, all tested bacterial species induced increased TREM-1 gene expression (Fig. 2B). In the case of S. pneumoniae at 24 h exposure, the bacteria caused very significant monocyte cytotoxicity. Hence, the RNA yield and quality from these samples was low, resulting in failure of qPCR at this time-point. Gram-positive organisms generally induced higher TREM-1 expression in THP-1 cells compared with the Gram-negative organisms at this time-point. Similar to TREM-1 gene expression, S. pneumoniae (MOI 100) induced significantly higher DAP12 expression following 4 h exposure (Fig. 2C). No other bacterial strain tested induced increased DAP12 expression at this time-point. However, after 24 h exposure, all bacterial strains tested, with the exception of T. forsythia, induced increased DAP12 expression in THP-1 cells (Fig. 2D).
Figure 2.
Relative expression of TREM-1 and DAP12 genes. THP-1 cells were exposed to bacteria at multiplicity of infection (MOI) of 10 or 100. After 4 and 24 h post exposure RNA was extracted, cDNA synthesized and the relative gene expression was determined by quantitative polymerase chain reaction, as described. (A) TREM-1 at 4 h; (B) TREM-1 at 24 h, (C) DAP12 at 4 h, (D) DAP12 at 24 h. Experiments were repeated three times. Data shown are mean ± SD of triplicates from a representative experiment. * P ≤ 0.05 compared with uninfected control.
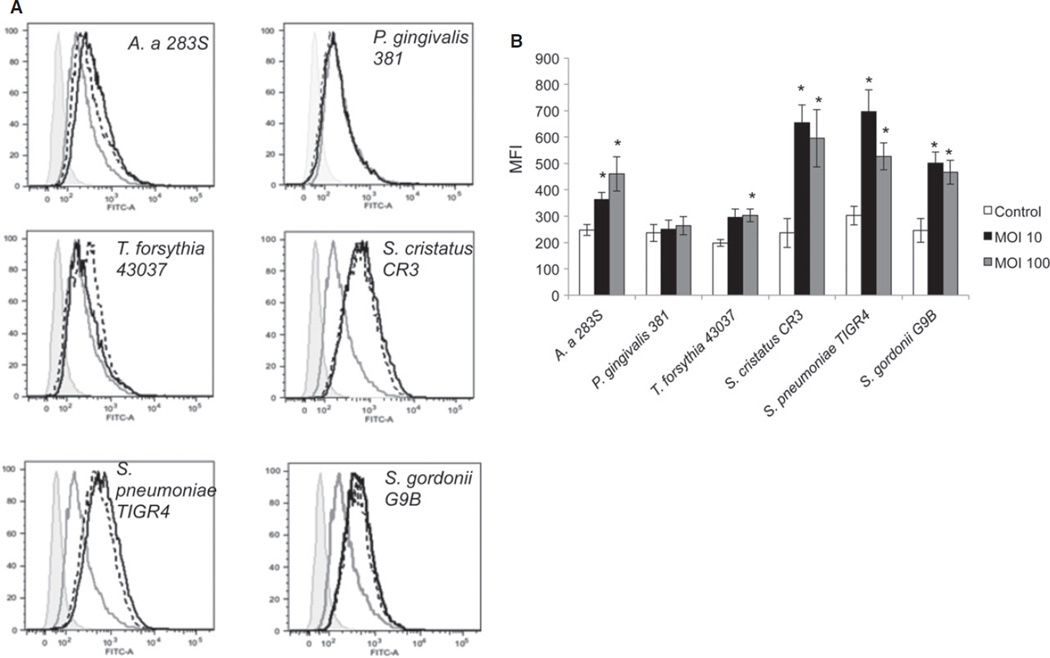
Surface expression of TREM-1
Changes in surface expression of TREM-1 were additionally examined (Fig. 3A,B). Exposure of THP-1 cells to bacteria for 24 h resulted in significantly increased surface expression of TREM-1 in all cases, with the exception of P. gingivalis. No differences in THP-1 surface expression of TREM-1 were seen after 4 h exposure to any of the tested bacteria (data not shown).
Figure 3.
Surface expression of TREM-1 receptors. THP-1 cells were exposed to the indicated oral bacteria at multiplicity of infection (MOI) of 10 and 100 for 24 h. Surface expression of TREM-1 receptor was determined by flow cytometry. (A) Histograms from a representative experiment, (grey filled – isotype control, grey line- uninfected controls, black dotted lines – MOI 10, black solid lines – MOI 100). (B) Mean fluorescence intensity (MFI) from two independent experiments for Tannerella forsythia and three independent experiments for all the other bacteria * P ≤ 0.05 compared with uninfected control.
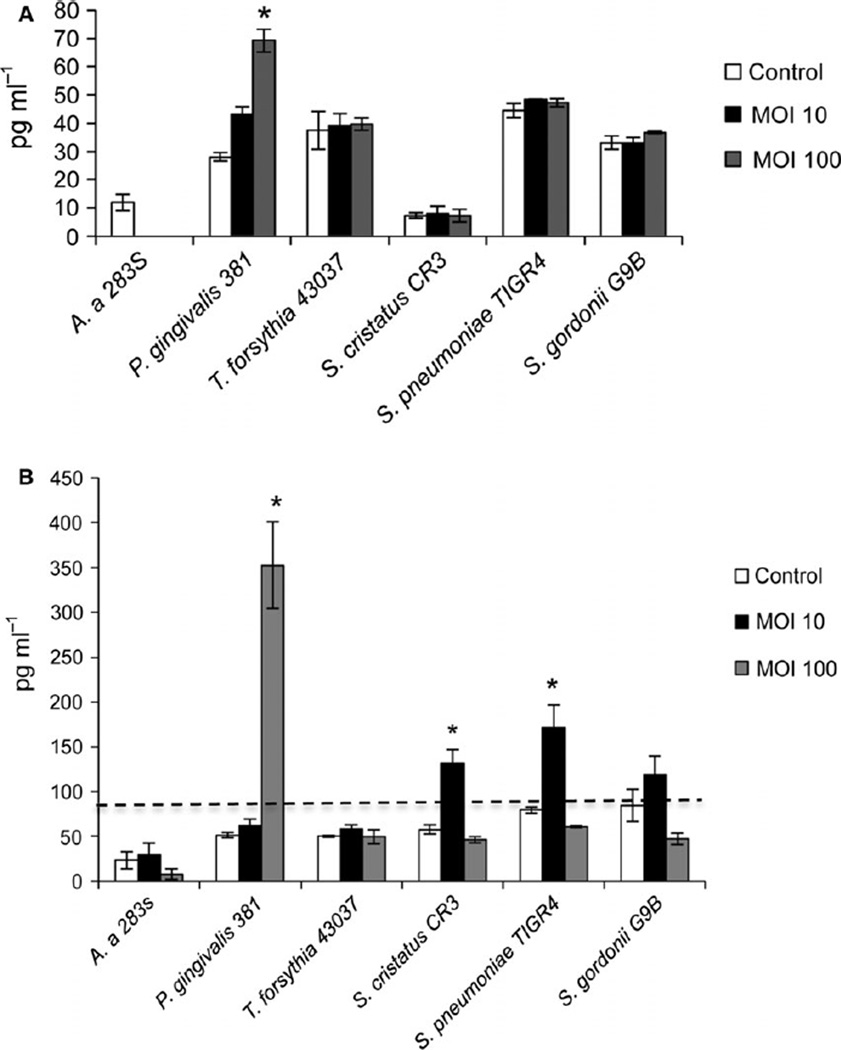
sTREM-1 release
The release of sTREM-1 in cell culture supernatants was determined by ELISA. Porphyromonas gingivalis (MOI 100) was the only bacterial strain to induce significant levels of sTREM-1 at 4 h (Fig. 4A), and at 24 h it induced the highest levels of sTREM-1 (Fig. 4B). Moderate levels of sTREM-1 were also induced by S. cristatus and S. pneumoniae at 24 h (MOI 10). sTREM-1 release was not induced by A. actinomycetemcomitans, T. forsythia or S. gordonii.
Figure 4.
Soluble TREM-1 (sTREM-1) release from THP-1 cells exposed to oral bacteria. THP-1 cells were exposed to bacteria at an MOI of 10 or 100. After 4 h (A) and 24 h (B) exposure, supernatants were collected and sTREM-1 levels were measured by enzyme-linked immunosorbent assay. Data shown are mean ± SD from three replicates. Line (- - -) indicates the start of the linear range of the assay (93.8 to 6000 pg ml−1). * P ≤ 0.05 compared with uninfected control.
Cytokine production and effect of TREM-1 inhibitor
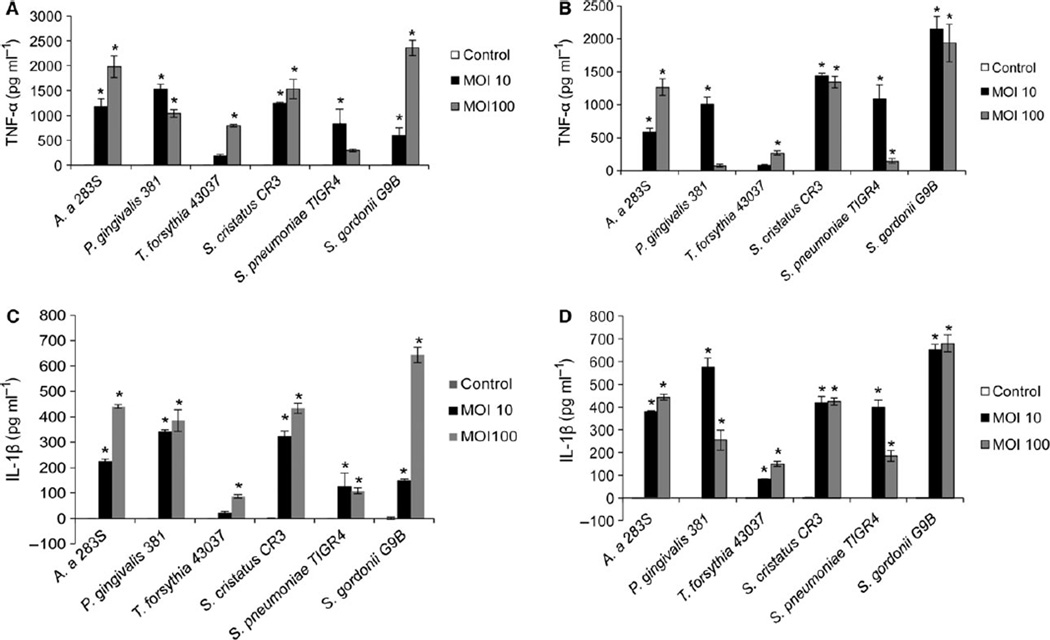
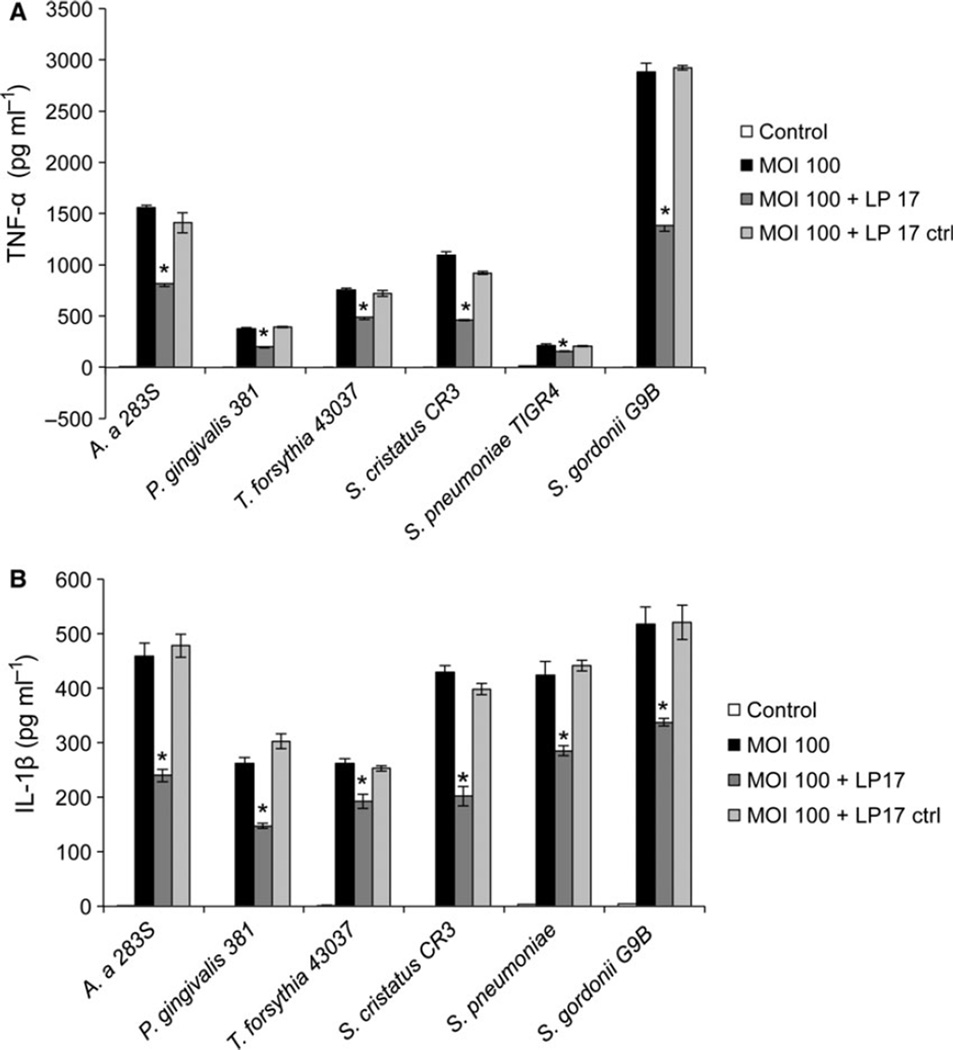
Exposure of THP-1 cells to oral bacteria resulted in proinflammatory cytokine production in all cases, as determined by measuring IL-1β and TNF-α release into the cell culture supernatant by ELISA (Fig. 5). We found the release of these inflammatory cytokines was significantly reduced when the THP-1 cells were incubated with the TREM-1 inhibitor LP-17 peptide simultaneously with the bacteria (Fig. 6). TNF-α was reduced 26–57% and IL-1β was reduced 26.4–53.2% by LP-17 treatment compared with control cells incubated with bacteria alone or with bacteria and a control peptide (LP-17 control peptide in Fig. 6).
Figure 5.
Pro-inflammatory cytokine release from THP-1 cells exposed to oral bacteria. THP-1 cells were exposed to the indicated oral bacteria at multiplicity of infection (MOI) of 10 and 100. After 4 and 24 h exposure supernatants were collected. Tumour necrosis factor-α (TNF-α) and interleukin-1 β (IL-1β) levels were estimated by enzyme-linked immunosorbent assay. (A) TNF-α at 4 h, (B) TNF-α at 24 h, (C) IL-1β at 4 h, (D) IL-1β at 24 h. Control is THP-1 cells only. Shown are means ± SD from three independent experiments. * P ≤ 0.05 compared with uninfected controls.
Figure 6.
Effect of TREM-1 inhibitor on cytokine production. THP-1 cells were exposed to indicated oral bacteria at a multiplicity of infection (MOI) of 100 for 24 h. Some of the wells were also treated with the LP17 or LP17 control peptide at a concentration of 100 ng ml−1. After 24 h the supernatant was collected and tumour necrosis factor-α (TNF-α) (A) and interleukin-1β (IL-1β) (B) levels were determined by enzyme-linked immunosorbent assay. Data shown are mean ± SD from three replicates. * P ≤ 0.05 compared with samples treated with bacteria only. Control is THP-1 cells only.
DISCUSSION
Periodontitis is the most prevalent disease of the oral cavity. The multispecies oral biofilm, along with the host immune response, are responsible for the onset and progression of this disease. The present study investigated TREM-1 production, as well as its importance in signaling for the production of proinflammatory cytokines, by human monocytes exposed to a variety of oral bacteria. It has been reported that P. gingivalis infection of the MonoMac-6 cell line resulted in the upregulation of TREM-1 and DAP12 genes, and was associated with increased sTREM levels in culture supernatants (Bostanci et al., 2011). This study confirms this finding, but additionally shows that TREM-1 is important for the immune reaction to Gram-positive commensal as well as Gram-negative pathogenic species.
Commensal bacteria play an important role in immunomodulation of tissues that support polymicrobial communities, such as the gut and oral cavity. Any imbalance in resident microbiota may lead to disease conditions such as periodontitis (Darveau, 2010) and inflammatory bowel disease (Serban, 2015). Commensal bacteria can also help to prime the immune system, enabling a rapid and efficient response to pathogens (Ivanov & Honda, 2012). Activation of the TREM-1 pathway by the commensals may help to sensitize the immune system to other oral pathogens. The normal healthy host regulates the inflammatory response to commensals through the mucosal immune mechanisms (Tlaskalova-Hogenova et al., 2004). Epithelial cells are an important component of the mucosal immune response, and depending upon the location they may be unresponsive to the common pathogen-associated molecular patterns, as shown by the reduced expression of TLR-4 in intestinal epithelial cells (Abreu et al., 2001). Also, it has been shown that intestinal macrophages do not express CD14 and CD89 (receptor for IgA) in healthy individuals and may contribute to the low level of inflammation in healthy intestinal mucosa (Smith et al., 2001). When the mucosal barrier is breached, however, the commensal bacteria may induce a proinflammatory response. Nakanishi and co-workers showed that breakdown of the intestinal epithelial barrier can result in an improper immune response to the Gram-positive intestinal commensal organisms leading to the development of colitis (Nakanishi et al., 2015). This may apply to the oral cavity, as expression of TLR-4 and CD14 vary within the oral cavity (Li et al., 2014). Although only Gram-positive oral commensals were included in the current study, we expect that both Gram-positive and Gram-negative bacteria once exposed to monocytes will induce a proinflammatory response.
TREM-1 is an amplifier of inflammation that depends on the association with the adaptor protein DAP12 for downstream signal transduction (Bouchon et al., 2000), which culminates in increased proinflammatory cytokine production. DAP12 is a transmembrane adapter protein that partners with a number of different cell surface receptors including TREM-1, TREM-2 and MDL-1 (Hamerman et al., 2009), leading to either activation or inhibition of cellular responses depending on the affinity/avidity of ligand binding (Hamerman et al., 2009). In the present study, as early as 4 h, P. gingivalis- and S. pneumoniae-exposed monocytes showed increased TREM-1 expression, and S. pneumoniae co-incubation resulted in significantly higher monocyte DAP12 expression as well. At 24 h post-exposure, both TREM-1 and DAP12 gene expression were upregulated in THP-1 monocytes by co-incubation with nearly all bacteria tested.
There are very few studies on the involvement of DAP12 in host response to bacterial pathogens. MonoMac-6 cells co-incubated with P. gingivalis W50 showed increased expression of DAP12 at MOIs of 1 and 10 after 24 h of infection, whereas there was no significant increase in DAP12 expression at an MOI of 100 (Bostanci et al., 2011). We found increased DAP12 expression both with MOI 10 and 100. Additionally, DAP12-deficient mice infected with S. pneumoniae have increased survival and reduced lung inflammation, concomitant with increased TNF-α production and phagocytic activation of alveolar macrophages (Hommes et al., 2014b). These results indicate that upregulation of DAP12 could be beneficial for the pathogens. In addition to TREM-1, DAP12 is associated with various other inhibitory and activating receptors including TREM-2, myeloid DAP12-associated lectin-1 (MDL-1) and signal-regulatory protein β1 (SIRPβ1) (Lanier, 2009). Whether interactions of pathogenic bacteria with TREM-1, or other interaction partners of DAP12, are more important for modifying the immune response to favor their survival remains unclear, though we did find inhibition of TREM-1 reduced the activation of THP-1 cells, suggesting the latter.
TREM-1 exists in two forms; as a surface receptor and a soluble peptide (sTREM-1) (Bouchon et al., 2001). sTREM-1 is believed to be generated either as a result of proteolytic cleavage of the surface receptor (Gomez-Pina et al., 2007) or by alternate splicing (Gingras et al., 2002; Baruah et al., 2015). Although sTREM cannot induce signal transduction, it likely competes with the surface receptor for available ligand, thereby acting as an antagonist to TREM-1 signaling. Porphyromonas gingivalis increases sTREM-1 through cleavage of the surface receptor by the bacterial protease enzymes called gingipains (Darveau, 2010). So while we did not find a decrease in the surface levels of TREM-1 below that seen in the uninfected controls, when combined with increased gene expression and significantly elevated sTREM-1 levels it suggests the possibility that cleavage of the surface TREM-1 is an additional mechanism by which P. gingivalis can suppress the local immune response (Darveau et al., 1998; Hajishengallis & Lamont, 2014). However, our data do not definitely rule out the possibility of alternate splicing. We also observed moderate amounts of sTREM-1 in supernatants from cells treated with S. pneumoniae and S. cristatus, consistent with reports of increased sTREM-1 levels in the bronchoalveolar lavage and lung homogenates of mice infected with S. pneumoniae (Hommes et al., 2014a). Further studies are required to determine if proteases produced by early colonizers such as S. cristatus (Wang et al., 2011) are capable of cleaving TREM-1 to actively modify the immune response.
Activation of TREM-1 also resulted in increased production of proinflammatory cytokines by the monocytic cells, as they were significantly reduced when the cells were treated with TREM-1 inhibitor. The inhibitory effect of the LP-17 peptide supports the notion that it competes with TREM-1 for the binding of the available ligand thereby inhibiting the TREM-1 pathway. The increase in cytokines is consistent with earlier reports that stimulation of TREM-1 can result in synergistic upregulation of signaling initiated by other pattern recognition receptors such as the TLRs (Arts et al., 2011, 2013; Prufer et al., 2014). Streptococcus pneumoniae induced lesser amounts of cytokines at MOI 100 when compared with the MOI 10. This may be because S. pneumoniae induced a very significant amount of cytotoxicity of THP-1 cells at MOI of 100. We also noticed a reduction in the cytokine production at MOI of 100 for P. gingivalis, known previously to cause degradation of released cytokines locally (Fletcher et al., 1997). Additionally, we found that treatment with the TREM-1 inhibitor did not abrogate cytokine production completely, as previously noted for other non-oral bacterial infections (Bostanci et al., 2011; Hu et al., 2014).
In summary, this study compared the activation of the TREM-1/DAP12 pathway by both commensal and pathogenic oral bacteria and found that both activate the pathway. The oral cavity supports a multispecies microbial community that stimulates inflammation as a default condition, similar to the situation seen in the gut (Devine et al., 2015), in which commensal bacteria play an important role regulating innate immune processes to maintain health. Future studies may investigate strategies using this information to diminish imbalance in the innate immune response to prevent or diminish inflammation and hence periodontal disease activity.
Acknowledgments
This study was supported by the NIDCR training grant 5T32DE023526. We thank Dr. Ashu Sharma for kindly providing Tannerella forsythia 43037 strain and for his valuable suggestions.
REFERENCES
- Abreu MT, Vora P, Faure E, Thomas LS, Arnold ET, Arditi M. Decreased expression of Toll-like receptor-4 and MD-2 correlates with intestinal epithelial cell protection against dysregulated proinflammatory gene expression in response to bacterial lipopolysaccharide. J Immunol. 2001;167:1609–1616. doi: 10.4049/jimmunol.167.3.1609. [DOI] [PubMed] [Google Scholar]
- Arts RJ, Joosten LA, Dinarello CA, Kullberg BJ, van der Meer JW, Netea MG. TREM-1 interaction with the LPS/TLR4 receptor complex. Eur Cytokine Netw. 2011;22:11–14. doi: 10.1684/ecn.2011.0274. [DOI] [PubMed] [Google Scholar]
- Arts RJ, Joosten LA, van der Meer JW, Netea MG. TREM-1: intracellular signaling pathways and interaction with pattern recognition receptors. J Leukoc Biol. 2013;93:209–215. doi: 10.1189/jlb.0312145. [DOI] [PubMed] [Google Scholar]
- Baruah S, Keck K, Vrenios M, et al. Identification of a novel splice variant isoform of TREM-1 in human neutrophil granules. J Immunol. 2015;195:5725–5731. doi: 10.4049/jimmunol.1402713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson C, Massin F, Lefevre PA, Thilly N, Miller N, Gibot S. Increased gingival crevicular fluid levels of soluble triggering receptor expressed on myeloid cells (sTREM) −1 in severe periodontitis. J Clin Periodontol. 2012;39:1141–1148. doi: 10.1111/jcpe.12008. [DOI] [PubMed] [Google Scholar]
- Bostanci N, Thurnheer T, Belibasakis GN. Involvement of the TREM-1/DAP12 pathway in the innate immune responses to Porphyromonas gingivalis. Mol Immunol. 2011;49:387–394. doi: 10.1016/j.molimm.2011.09.012. [DOI] [PubMed] [Google Scholar]
- Bouchon A, Dietrich J, Colonna M. Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J Immunol. 2000;164:4991–4995. doi: 10.4049/jimmunol.164.10.4991. [DOI] [PubMed] [Google Scholar]
- Bouchon A, Facchetti F, Weigand MA, Colonna M. TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature. 2001;410:1103–1107. doi: 10.1038/35074114. [DOI] [PubMed] [Google Scholar]
- Buckland KF, Ramaprakash H, Murray LA, et al. Triggering receptor expressed on myeloid cells-1 (TREM-1) modulates immune responses to Aspergillus fumigatus during fungal asthma in mice. Immunol Invest. 2011;40:692–722. doi: 10.3109/08820139.2011.578270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- Darveau RP, Belton CM, Reife RA, Lamont RJ. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect Immun. 1998;66:1660–1665. doi: 10.1128/iai.66.4.1660-1665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine DA, Marsh PD, Meade J. Modulation of host responses by oral commensal bacteria. J Oral Microbiol. 2015;7:26941. doi: 10.3402/jom.v7.26941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eke PI, Dye BA, Wei L, Thornton-Evans GO, Genco RJ. Prevalence of periodontitis in adults in the United States: 2009 and 2010. J Dent Res. 2012;91:914–920. doi: 10.1177/0022034512457373. [DOI] [PubMed] [Google Scholar]
- El Mezayen R, El Gazzar M, Seeds MC, McCall CE, Dreskin SC, Nicolls MR. Endogenous signals released from necrotic cells augment inflammatory responses to bacterial endotoxin. Immunol Left. 2007;111:36–44. doi: 10.1016/j.imlet.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J, Reddi K, Poole S, et al. Interactions between periodontopathogenic bacteria and cytokines. J Periodontal Res. 1997;32:200–205. doi: 10.1111/j.1600-0765.1997.tb01406.x. [DOI] [PubMed] [Google Scholar]
- Gibot S, Kolopp-Sarda MN, Bene MC, et al. A soluble form of the triggering receptor expressed on myeloid cells-1 modulates the inflammatory response in murine sepsis. J Exp Med. 2004;200:1419–1426. doi: 10.1084/jem.20040708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibot S, Massin F, Marcou M, et al. TREM-1 promotes survival during septic shock in mice. Eur J Immunol. 2007;37:456–466. doi: 10.1002/eji.200636387. [DOI] [PubMed] [Google Scholar]
- Gingras MC, Lapillonne H, Margolin JF. TREM-1, MDL-1, and DAP12 expression is associated with a mature stage of myeloid development. Mol Immunol. 2002;38:817–824. doi: 10.1016/s0161-5890(02)00004-4. [DOI] [PubMed] [Google Scholar]
- Gomez-Pina V, Soares-Schanoski A, Rodriguez-Rojas A, et al. Metalloproteinases shed TREM-1 ectodomain from lipopolysaccharide-stimulated human monocytes. J Immunol. 2007;179:4065–4073. doi: 10.4049/jimmunol.179.6.4065. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G, Lamont RJ. Breaking bad: manipulation of the host response by Porphyromonas gingivalis. Eur J Immunol. 2014;44:328–338. doi: 10.1002/eji.201344202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamerman JA, Ni M, Killebrew JR, Chu CL, Lowell CA. The expanding roles of ITAM adapters FcRγ and DAP12 in myeloid cells. Immunol Rev. 2009;232:42–58. doi: 10.1111/j.1600-065X.2009.00841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselmayer P, Grosse-Hovest L, von Landenberg P, Schild H, Radsak MP. TREM-1 ligand expression on platelets enhances neutrophil activation. Blood. 2007;110:1029–1035. doi: 10.1182/blood-2007-01-069195. [DOI] [PubMed] [Google Scholar]
- Haubek D, Ennibi OK, Poulsen K, Vaeth M, Poulsen S, Kilian M. Risk of aggressive periodontitis in adolescent carriers of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans in Morocco: a prospective longitudinal cohort study. Lancet. 2008;371:237–242. doi: 10.1016/S0140-6736(08)60135-X. [DOI] [PubMed] [Google Scholar]
- Hommes TJ, Hoogendijk AJ, Dessing MC, et al. Triggering receptor expressed on myeloid cells-1 (TREM-1) improves host defence in pneumococcal pneumonia. J Pathol. 2014a;233:357–367. doi: 10.1002/path.4361. [DOI] [PubMed] [Google Scholar]
- Hommes TJ, Hoogendijk AJ, Dessing MC, et al. DNAX-activating protein of 12 kDa impairs host defense in pneumococcal pneumonia. Crit Care Med. 2014b;42:e783–e790. doi: 10.1097/CCM.0000000000000686. [DOI] [PubMed] [Google Scholar]
- Hu LT, Du ZD, Zhao GQ, et al. Role of TREM-1 in response to Aspergillus fumigatus infection in corneal epithelial cells. Int Immunopharmacol. 2014;23:288–293. doi: 10.1016/j.intimp.2014.09.011. [DOI] [PubMed] [Google Scholar]
- Ivan FX, Rajapakse JC, Welsch RE, et al. Differential pulmonary transcriptomic profiles in murine lungs infected with low and highly virulent influenza H3N2 viruses reveal dysregulation of TREM1 signaling, cytokines, and chemokines. Fund Integr Genomics. 2012;12:105–117. doi: 10.1007/s10142-011-0247-y. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Honda K. Intestinal commensal microbes as immune modulators. Cell Host Microbe. 2012;12:496–508. doi: 10.1016/j.chom.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagler H, Sharif O, Haslinger I, et al. TREM-1 activation alters the dynamics of pulmonary IRAK-M expression in vivo and improves host defense during pneumococcal pneumonia. J Immunol. 2009;183:2027–2036. doi: 10.4049/jimmunol.0803862. [DOI] [PubMed] [Google Scholar]
- Lanier LL. DAP10- and DAP12-associated receptors in innate immunity. Immunol Rev. 2009;227:150–160. doi: 10.1111/j.1600-065X.2008.00720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JP, Chen Y, Ng CH, et al. Differential expression of Toll-like receptor 4 in healthy and diseased human gingiva. J Periodontal Res. 2014;49:845–854. doi: 10.1111/jre.12173. [DOI] [PubMed] [Google Scholar]
- Nakanishi Y, Sato T, Ohteki T. Commensal Gram-positive bacteria initiates colitis by inducing monocyte/macrophage mobilization. Mucosal Immunol. 2015;8:152–160. doi: 10.1038/mi.2014.53. [DOI] [PubMed] [Google Scholar]
- Oliveira RR, Fermiano D, Feres M, et al. Levels of candidate periodontal pathogens in subgingival biofilm. J Dent Res. 2016;95:711–718. doi: 10.1177/0022034516634619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornatowska M, Azim AC, Wang X, et al. Functional genomics of silencing TREM-1 on TLR4 signaling in macrophages. Am J Physiol Lung Cell Mol Physiol. 2007;293:L1377–L1384. doi: 10.1152/ajplung.00140.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo SJ, Simpson TA, Simmons JM, Schnapp LM. Soluble triggering receptor expressed on myeloid cells-1 (sTREM-1) as a diagnostic marker of ventilator-associated pneumonia. Respir Care. 2012;57:2052–2058. doi: 10.4187/respcare.01703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prufer S, Weber M, Sasca D, et al. Distinct signaling cascades of TREM-1, TLR and NLR in neutrophils and monocytic cells. J Innate Immun. 2014;6:339–352. doi: 10.1159/000355892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radaev S, Kattah M, Rostro B, Colonna M, Sun PD. Crystal structure of the human myeloid cell activating receptor TREM-1. Structured: 2003:1527–1535. doi: 10.1016/j.str.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Radsak MP, Salih HR, Rammensee HG, Schild H. Triggering receptor expressed on myeloid cells-1 in neutrophil inflammatory responses: differential regulation of activation and survival. J Immunol. 2004;172:4956–4963. doi: 10.4049/jimmunol.172.8.4956. [DOI] [PubMed] [Google Scholar]
- Ramanathan B, Minton JE, Ross CR, Blecha F. Cloning of porcine triggering receptor expressed on myeloid cells-1 (TREM-1) and its induction by lipopolysaccharide, peptidoglycan, and Salmonella enterica serovar Typhimurium infection. Dev Comp Immunol. 2005;29:1–7. doi: 10.1016/j.dci.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Read CB, Kuijper JL, Hjorth SA, et al. Cutting edge: identification of neutrophil PGLYRP1 as a ligand for TREM-1. J Immunol. 2015;194:1417–1421. doi: 10.4049/jimmunol.1402303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serban DE. Microbiota in inflammatory bowel disease pathogenesis and therapy: Is it all about diet? Nutr Clin Pract. 2015;30:760–779. doi: 10.1177/0884533615606898. [DOI] [PubMed] [Google Scholar]
- Sharif O, Knapp S. From expression to signaling: roles of TREM-1 and TREM-2 in innate immunity and bacterial infection. Immunobiology. 2008;213:701–713. doi: 10.1016/j.imbio.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Sharif O, Bolshakov VN, Raines S, Newham P, Perkins ND. Transcriptional profiling of the LPS induced NF-κB response in macrophages. BMC Immunol. 2007;8:1. doi: 10.1186/1471-2172-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PD, Smythies LE, Mosteller-Barnum M, et al. Intestinal macrophages lack CD14 and CD89 and consequently are down-regulated for LPS- and IgA-mediated activities. J Immunol. 2001;167:2651–2656. doi: 10.4049/jimmunol.167.5.2651. [DOI] [PubMed] [Google Scholar]
- Thomas P, Sekhar AC, Mujawar MM. Non-recovery of varying proportions of viable bacteria during spread plating governed by the extent of spreader usage and proposal for an alternate spotting-spreading approach to maximize the CFU. J Appl Microbiol. 2012;113:339–350. doi: 10.1111/j.1365-2672.2012.05327.x. [DOI] [PubMed] [Google Scholar]
- Tlaskalova-Hogenova H, Stepankova R, Hudcovic T, et al. Commensal bacteria (normal microflora), mucosal immunity and chronic inflammatory and autoimmune diseases. Immunol Lett. 2004;93:97–108. doi: 10.1016/j.imlet.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Wang BY, Deutch A, Hong J, Kuramitsu HK. Proteases of an early colonizer can hinder Streptococcus mutans colonization in vitro. J Dent Res. 2011;90:501–505. doi: 10.1177/0022034510388808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yucel-Lindberg T, Bage T. Inflammatory mediators in the pathogenesis of periodontitis. Expert Rev Mol Med. 2013;15:e7. doi: 10.1017/erm.2013.8. [DOI] [PubMed] [Google Scholar]