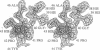Abstract
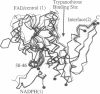
Trypanosomes and related protozoan parasites lack glutathione reductase and possess instead a closely related enzyme that serves as the reductant of a bis(glutathione)-spermidine conjugate, trypanothione. The human and parasite enzymes have mutually exclusive substrate specificities, providing a route for the design of therapeutic agents by specific inhibition of the parasite enzyme. We report here the three-dimensional structure of trypanothione reductase from Crithidia fasciculata and show that it closely resembles the structure of human glutathione reductase. In particular, the core structure surrounding the catalytic machinery is almost identical in the two enzymes. However, significant differences are found at the substrate binding sites. A cluster of basic residues in glutathione reductase is replaced by neutral, hydrophobic, or acidic residues in trypanothione reductase, consistent with the nature of the spermidine linkage and the change in overall charge of the substrate from -2 to +1, respectively. The binding site is more open in trypanothione reductase due to rotations of about 4 degrees in the domains that form the site, with relative shifts of as much as 2-3 A in residue positions. These results provide a detailed view of the residues that can interact with potential inhibitors and complement previous modeling and mutagenesis studies on the two enzymes.
Full text
PDF
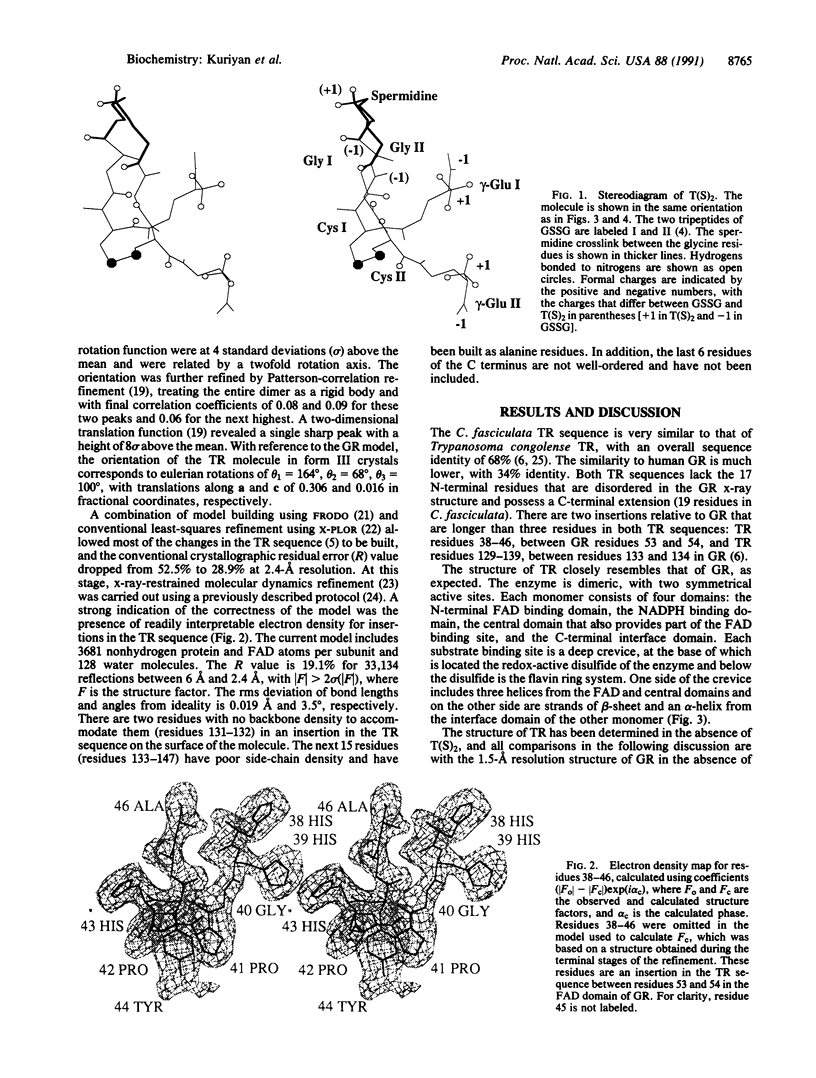

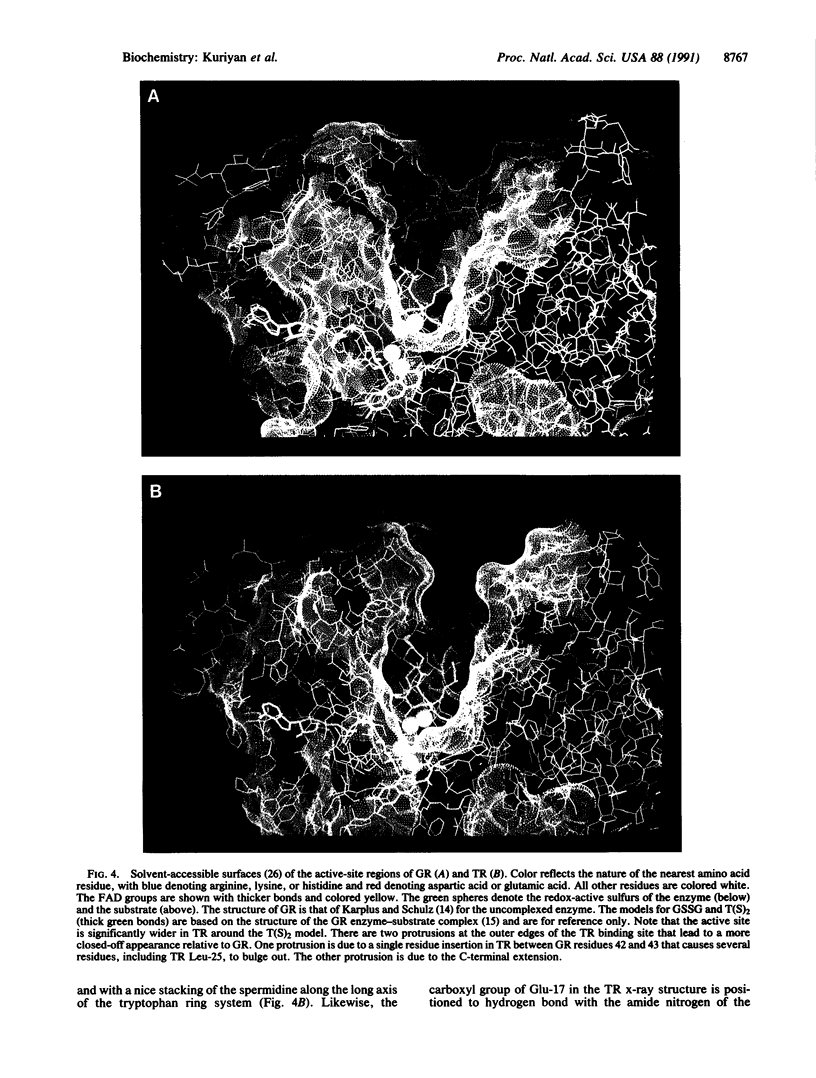

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arrick B. A., Griffith O. W., Cerami A. Inhibition of glutathione synthesis as a chemotherapeutic strategy for trypanosomiasis. J Exp Med. 1981 Mar 1;153(3):720–725. doi: 10.1084/jem.153.3.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Fairlamb A. H., Blackburn P., Ulrich P., Chait B. T., Cerami A. Trypanothione: a novel bis(glutathionyl)spermidine cofactor for glutathione reductase in trypanosomatids. Science. 1985 Mar 22;227(4693):1485–1487. doi: 10.1126/science.3883489. [DOI] [PubMed] [Google Scholar]
- Henderson G. B., Fairlamb A. H., Ulrich P., Cerami A. Substrate specificity of the flavoprotein trypanothione disulfide reductase from Crithidia fasciculata. Biochemistry. 1987 Jun 2;26(11):3023–3027. doi: 10.1021/bi00385a011. [DOI] [PubMed] [Google Scholar]
- Henderson G. B., Murgolo N. J., Kuriyan J., Osapay K., Kominos D., Berry A., Scrutton N. S., Hinchliffe N. W., Perham R. N., Cerami A. Engineering the substrate specificity of glutathione reductase toward that of trypanothione reduction. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8769–8773. doi: 10.1073/pnas.88.19.8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter W. N., Smith K., Derewenda Z., Harrop S. J., Habash J., Islam M. S., Helliwell J. R., Fairlamb A. H. Initiating a crystallographic study of trypanothione reductase. J Mol Biol. 1990 Nov 20;216(2):235–237. doi: 10.1016/S0022-2836(05)80314-6. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Thirup S. Using known substructures in protein model building and crystallography. EMBO J. 1986 Apr;5(4):819–822. doi: 10.1002/j.1460-2075.1986.tb04287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus P. A., Pai E. F., Schulz G. E. A crystallographic study of the glutathione binding site of glutathione reductase at 0.3-nm resolution. Eur J Biochem. 1989 Jan 2;178(3):693–703. doi: 10.1111/j.1432-1033.1989.tb14500.x. [DOI] [PubMed] [Google Scholar]
- Karplus P. A., Schulz G. E. Refined structure of glutathione reductase at 1.54 A resolution. J Mol Biol. 1987 Jun 5;195(3):701–729. doi: 10.1016/0022-2836(87)90191-4. [DOI] [PubMed] [Google Scholar]
- Karplus P. A., Schulz G. E. Substrate binding and catalysis by glutathione reductase as derived from refined enzyme: substrate crystal structures at 2 A resolution. J Mol Biol. 1989 Nov 5;210(1):163–180. doi: 10.1016/0022-2836(89)90298-2. [DOI] [PubMed] [Google Scholar]
- Krauth-Siegel R. L., Enders B., Henderson G. B., Fairlamb A. H., Schirmer R. H. Trypanothione reductase from Trypanosoma cruzi. Purification and characterization of the crystalline enzyme. Eur J Biochem. 1987 Apr 1;164(1):123–128. doi: 10.1111/j.1432-1033.1987.tb11002.x. [DOI] [PubMed] [Google Scholar]
- Kuriyan J., Wong L., Guenther B. D., Murgolo N. J., Cerami A., Henderson G. B. Preliminary crystallographic analysis of trypanothione reductase from Crithidia fasciculata. J Mol Biol. 1990 Oct 5;215(3):335–337. doi: 10.1016/s0022-2836(05)80353-5. [DOI] [PubMed] [Google Scholar]
- Lee B., Richards F. M. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971 Feb 14;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Meshnick S. R., Blobstein S. H., Grady R. W., Cerami A. An approach to the development of new drugs for African trypanosomiasis. J Exp Med. 1978 Aug 1;148(2):569–579. doi: 10.1084/jem.148.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgolo N. J., Cerami A., Henderson G. B. Biomedical science and the third world. Under the volcano. Trypanothione reductase. Ann N Y Acad Sci. 1989;569:193–200. doi: 10.1111/j.1749-6632.1989.tb27369.x. [DOI] [PubMed] [Google Scholar]
- Shames S. L., Fairlamb A. H., Cerami A., Walsh C. T. Purification and characterization of trypanothione reductase from Crithidia fasciculata, a newly discovered member of the family of disulfide-containing flavoprotein reductases. Biochemistry. 1986 Jun 17;25(12):3519–3526. doi: 10.1021/bi00360a007. [DOI] [PubMed] [Google Scholar]
- Shames S. L., Kimmel B. E., Peoples O. P., Agabian N., Walsh C. T. Trypanothione reductase of Trypanosoma congolense: gene isolation, primary sequence determination, and comparison to glutathione reductase. Biochemistry. 1988 Jul 12;27(14):5014–5019. doi: 10.1021/bi00414a010. [DOI] [PubMed] [Google Scholar]
- Sullivan F. X., Sobolov S. B., Bradley M., Walsh C. T. Mutational analysis of parasite trypanothione reductase: acquisition of glutathione reductase activity in a triple mutant. Biochemistry. 1991 Mar 19;30(11):2761–2767. doi: 10.1021/bi00225a004. [DOI] [PubMed] [Google Scholar]
- Sullivan F. X., Walsh C. T. Cloning, sequencing, overproduction and purification of trypanothione reductase from Trypanosoma cruzi. Mol Biochem Parasitol. 1991 Jan;44(1):145–147. doi: 10.1016/0166-6851(91)90231-t. [DOI] [PubMed] [Google Scholar]
- Weis W. I., Brünger A. T., Skehel J. J., Wiley D. C. Refinement of the influenza virus hemagglutinin by simulated annealing. J Mol Biol. 1990 Apr 20;212(4):737–761. doi: 10.1016/0022-2836(90)90234-D. [DOI] [PubMed] [Google Scholar]