Figure 6.
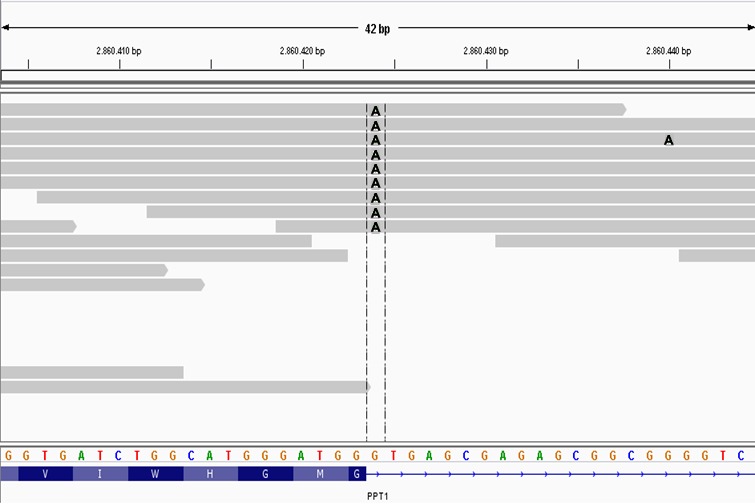
A screen capture of the affected Cane Corso's whole genome sequence alignment over 42 bp of canine chromosome 15 visualized with the Integrative Genomics Viewer. Each gray bar represents sequence reads aligned to the CanFam3.1 reference sequence assembly shown with the 4‐color nucleotide sequence near the bottom of the figure. Variants in the sequence reads are indicated by letters at the pertinent positions within the gray bars. In this case, the As within the gray bars indicate the variant's position where an A in a sequence read replaces a G from the reference sequence. Colors were modified from the screen capture to enhance visibility of the As. The track at the bottom shows the amino acid sequence translated from codons at the 3′ end of PPT1 exon 1. The horizontal dashed lines enclose the homozygous As at position +1 of intron 1 that destroy the canonical GT splice donor motif.
