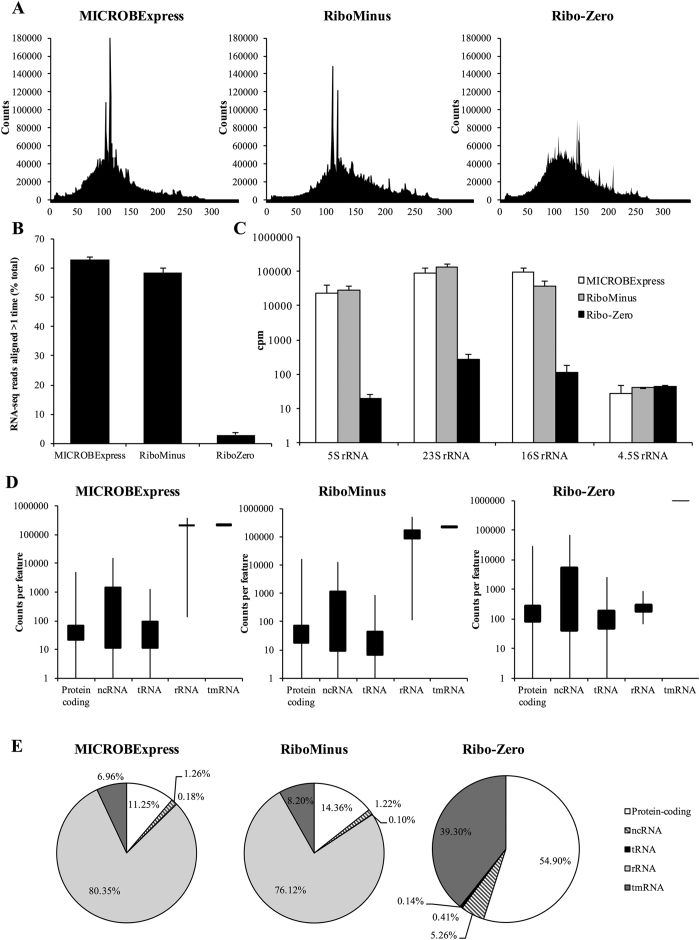
Figure 3. Ribo-Zero treatment reduces rRNA reads and increases non-rRNA reads during RNA-seq analysis.
(A) Read length histogram from RNA-seq analysis of MICROBExpress-, RiboMinus-, and Ribo-Zero-treated samples performed on the Ion Torrent PGM system using the Ion PGM Template OT2 200 and Ion PGM Sequencing 200 v2 kits. Representative histogram from one RNA-seq run is shown. (B) Percentage of RNA-seq reads that aligned to more than one location on the P. aeruginosa PAO1 genome during bowtie-2 read mapping. (C) Counts per million (cpm) of detected reads matching rRNA-coding elements in the indicated RNA-sequencing samples, as determined by computing counts using Qualimap. Cpm, counts per million. Error bars indicate standard deviation. (D) Average number of counts detected per biotype for the indicated RNA-seq samples as determined using the NOISeq package software. (E) Counts detected per biotype as a percentage of total counts of the indicated RNA-seq runs. Data were derived from two independent RNA-seq experiments using biological replicates.