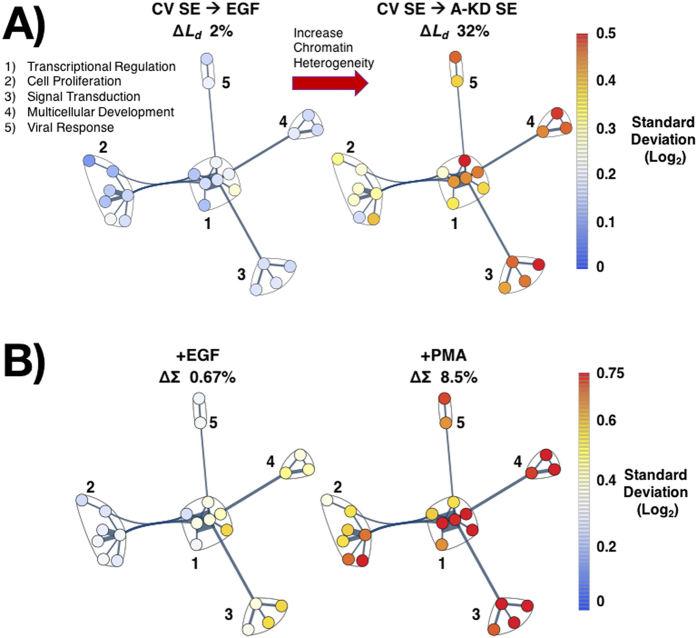
Figure 3. Transcriptional network heterogeneity increases with changes in the physical topology of the chromatin nanoarchitecture.
(A,B) Analysis of cluster domains for 22 GO processes that contain at least 10 genes. Each point represents a GO ontological process organized by a spring-electrical distribution of ontologies. Ontologies that are highly interconnected with respect to the number of shared genes self-organize into functional domains representing: (1) Transcriptional Regulation, (2) Signal Transduction, (3) Multicellular Development, (4) Viral Response, and (5) Cellular Proliferation. Ontologies are pseudo-colored based on their intra-network heterogeneity calculated as the standard deviation of relative expression for genes,  , belonging to that process, P, between indicated conditions: Het(P) = Standard Deviation
, belonging to that process, P, between indicated conditions: Het(P) = Standard Deviation 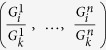 , where k indicates the reference condition and i the comparison condition. For full calculation, see Equation (6). (A) Intra-network heterogeneity of gene expression for the ontologies described above using CV SE as the reference condition in comparison to CV EGF (ΔLd = 2%, the left graph) and in comparison to A-KD SE (ΔLd = 32%, the right graph). Chromatin heterogeneity Ld was measured in fixed cells. A higher ΔLd between conditions is associated with increased divergence of gene expression within any given process. (B) Analysis of ontological divergence as described above in relation to changes in chromatin heterogeneity in live cells measured in real-time. Transformation of intra-network expression is analyzed relative to the CV SD (initial state) as the reference condition to compare the transformation for two final states: + EGF (Δ∑ = 0.67%, the right graph) and +PMA (Δ∑ = 8.5%, the left graph). Chromatin heterogeneity Δ∑ was measured in the same live cells before and after treatment. Early transformation in chromatin topology, Δ∑, precedes observed intra-network transcriptional heterogeneity measured through microarray analysis. Critically, measurements of ∑ were taken within 30 minutes, timescales which precede the classical expectation of intra-network feedback mechanisms due to translational feedback.
, where k indicates the reference condition and i the comparison condition. For full calculation, see Equation (6). (A) Intra-network heterogeneity of gene expression for the ontologies described above using CV SE as the reference condition in comparison to CV EGF (ΔLd = 2%, the left graph) and in comparison to A-KD SE (ΔLd = 32%, the right graph). Chromatin heterogeneity Ld was measured in fixed cells. A higher ΔLd between conditions is associated with increased divergence of gene expression within any given process. (B) Analysis of ontological divergence as described above in relation to changes in chromatin heterogeneity in live cells measured in real-time. Transformation of intra-network expression is analyzed relative to the CV SD (initial state) as the reference condition to compare the transformation for two final states: + EGF (Δ∑ = 0.67%, the right graph) and +PMA (Δ∑ = 8.5%, the left graph). Chromatin heterogeneity Δ∑ was measured in the same live cells before and after treatment. Early transformation in chromatin topology, Δ∑, precedes observed intra-network transcriptional heterogeneity measured through microarray analysis. Critically, measurements of ∑ were taken within 30 minutes, timescales which precede the classical expectation of intra-network feedback mechanisms due to translational feedback.