Fig. 1.
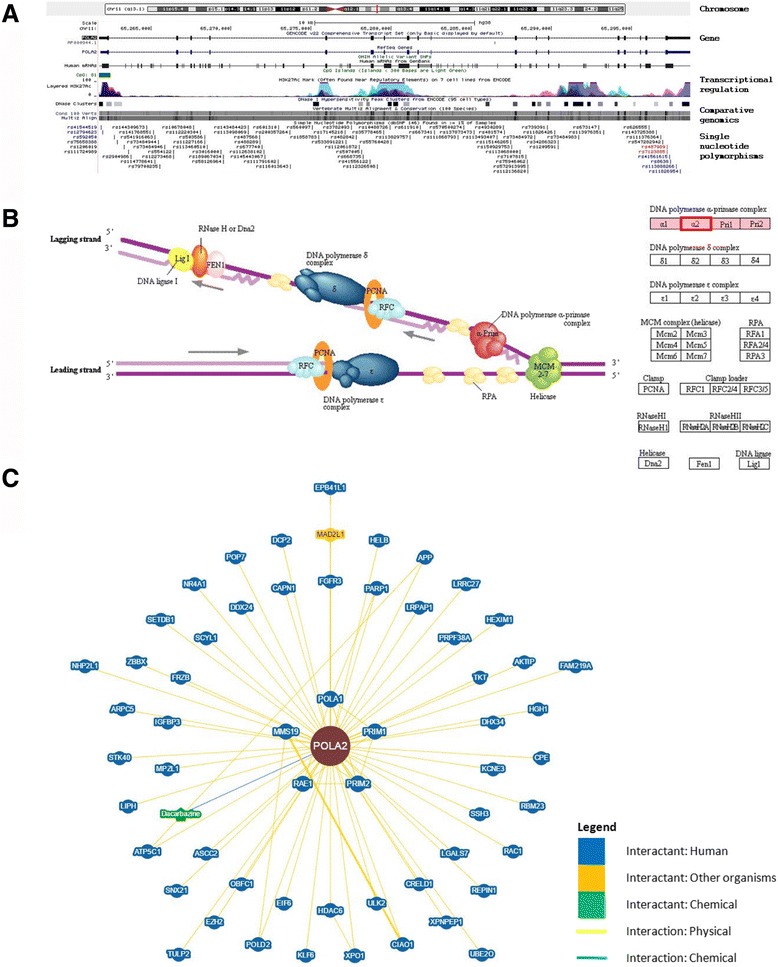
Genomics analysis of POLA2. a Genomic landscape of POLA2 based on the human Dec. 2013 (GRCh38/hg38) assembly. Location of POLA2 is indicated by a red vertical line on chromosome 11. Coding exons are represented by blocks linked by a horizontal line. CpG islands are shown as green blocks. Data associated with integrated transcriptional regulatory elements were retrieved from ENCODE. Comparative genomics analysis using multiple alignments of vertebrate species revealed conservation of the POLA2 gene. Each SNP is shown separately and labeled with the respective SNP ID reported by dbSNP build 146. Only SNPs that have a minor allele frequency of at least 1% and are mapped to a single location in the reference genome assembly are shown in the figure. b Eukaryotic DNA replication complex showing the role of POLA2 (boxed in red) in DNA replication, which involves a complex network of interacting enzymes and proteins. Three DNA polymerases (α, δ and ε) have been identified in eukaryotes. DNA polymerase α, including POLA2, forms a complex with DNA primase during the process. Source: Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database. c Network visualization of POLA2 and its interactants. Physical and genetic interactions, including chemical associations and post-translational modifications, were curated from published datasets and annotated interaction data from NCBI to construct this network
