Abstract
Aim:
Phosphodiesterase 4 (PDE4) isozymes are involved in different functions, depending on their patterns of distribution in the brain. The PDE4 subtypes are distributed in different inflammatory cells, and appear to be important regulators of inflammatory processes. In this study we examined the effects of ferulic acid (FA), a plant component with strong anti-oxidant and anti-inflammatory activities, on lipopolysaccharide (LPS)-induced up-regulation of phosphodiesterase 4B (PDE4B) in PC12 cells, which in turn regulated cellular cAMP levels and the cAMP/cAMP response element binding protein (CREB) pathway in the cells.
Methods:
PC12 cells were treated with LPS (1 μg/mL) for 8 h, and the changes of F-actin were detected using laser scanning confocal microscopy. The levels of pro-inflammatory cytokines were measured suing ELISA kits, and PDE4B-specific enzymatic activity was assessed with a PDE4B assay kit. The mRNA levels of PDE4B were analyzed with Q-PCR, and the protein levels of CREB and phosphorylated CREB (pCREB) were determined using immunoblotting. Furthermore, molecular docking was used to identify the interaction between PDE4B2 and FA.
Results:
Treatment of PC12 cells with LPS induced thick bundles of actin filaments appearing in the F-actin cytoskeleton, which were ameliorated by pretreatment with FA (10–40 μmol/L) or with a PDE4B inhibitor rolipram (30 μmol/L). Pretreatment with FA dose-dependently inhibited the LPS-induced production of TNF-α and IL-1β in PC12 cells. Furthermore, pretreatment with FA dose-dependently attenuated the LPS-induced up-regulation of PDE4 activity in PC12 cells. Moreover, pretreatment with FA decreased LPS-induced up-regulation of the PDE4B mRNA, and reversed LPS-induced down-regulation of CREB and pCREB in PC12 cells. The molecular docking results revealed electrostatic and hydrophobic interactions between FA and PDE4B2.
Conclusion:
The beneficial effects of FA in PC12 cells might be conferred through inhibition of LPS-induced up-regulation of PDE4B and stimulation of cAMP/CREB signaling pathway. Therefore, FA may be a potential therapeutic intervention for the treatment of neuroinflammatory diseases such as AD.
Keywords: ferulic acid, rolipram, PC12 cells, lipopolysaccharide, neuroinflammation, F-actin, TNF-α, IL-1β, PDE4B, CREB
Introduction
Neuro-inflammation is associated with a variety of neurodegenerative diseases such as Alzheimer's, Parkinson's and Huntington's diseases1. Lipopolysaccharide (LPS) is a potent stimulator of microglia that results in production of pro-inflammatory cytokines in the brain, and it has been widely used to induce inflammation in the brain and thereby contribute to neurodegeneration2,3. PC12 cells are a well-known model for studying neuronal signaling pathways and other neuro-biochemical events because they display phenotypic characteristics of adrenal chromaffin cells and sympathetic neurons4,5.
LPS is a typical inducer for PDE4B production and inflammatory factors, and LPS-induced brain inflammation results in the impairment of memory function and learning ability6,7,8,9. Studies by Reyes-Irisarri et al have demonstrated that LPS administration increases PDE4B2 mRNA expression in the choroid plexus10. PDE4 inhibitors suppress the LPS-induced TNF-α production in inflammation models including liver injury, lung injury, renal failure and sclerosis11,12.
The PDE4 subtype PDE4B is highly expressed in inflammatory, immune, and airway smooth muscle cells and is believed to play an important role in inflammation11,13. Substantial evidence has shown that PDE4B is highly expressed in the amygdala, striatum and hypothalamus, thus suggesting its potential for use in the treatment of anxiety and AD10,14,15,16,17. PDE4B is associated with long-term potentiation (LTP) in hippocampal neurons, thereby linking neuro-protection, anti-inflammation and PDE4B activity18,19,20.
Likewise, drugs that lower PDE4B expression and stimulate the cyclic AMP (cAMP)/cAMP response element binding protein (CREB) pathway may enhance anti-inflammatory activity. A number of antioxidants including ferulic acid (FA) and related ester derivatives decrease the levels of some inflammatory mediators, such as TNF-α and IL-1β, as well as limit their associated functions21,22,23.
FA is a plant component that is a free radical scavenger, and it exhibits strong anti-oxidant and anti-inflammatory capacity24. FA can easily cross the blood-brain barrier, and it exhibits many biological activities, thus potentially making it a useful agent for preventing neuro-inflammatory diseases25. Food supplementation with curcumin and ferulic acid is considered to be a nutritional approach to reduce oxidative damage and amyloid pathology in Alzheimer's, Parkinson's and Huntington's diseases26. The mRNA gene expression profiles indicate that FA mediates neuroprotection by down-regulating ROS and inflammatory and apoptotic markers. Ferulic acid effectively protects against further secondary impairments associated with traumatic brain injury27. Although FA has diverse biological functions, such as anti-inflammation, anti-cancer, anti-diabetes, and anti-atherosclerosis and neuroprotection against oxidative stress-related apoptosis28,29,30,31,32, its biological activities in the central nervous system (CNS) remain largely unknown. In the present study, we examined the effects of FA on the LPS-induced up-regulation of the expression and activity of PDE4B, which regulates cellular cyclic adenosine monophosphate (cAMP) levels and the cAMP/cAMP response element binding protein (CREB) pathway. Our previous studies have shown that FA increases cAMP levels and inhibits the LPS-induced increase in fluorescent intensity of Ca2+ in PC12 cells33. The stimulation of cAMP26 and inhibition of Ca2+ by FA is associated with decreased activity and expression of PDE4B. The recently reported cellular activities of FA may involve modulation of transcriptional factors such as CREB, which controls the expression of various genes implicated in inflammation, cell differentiation and proliferation34. It is of interest to examine how FA affects the production of PDE4B in response to LPS stimulation and thus exerts its neuroprotective effects. Our previous studies have also shown that FA inhibits the production of IL-1β and IL-TNF-α and blocks Ca2+ channels in PC12 cells33.
In the present work, LPS-induced PC12 cells were used to investigate the possible mechanism of neuroprotection by FA. We postulated that this anti-inflammatory activity of FA is likely to involve inhibition of LPS-induced PDE4B up-regulation and stimulation of the cAMP/CREB pathway.
Materials and methods
Cell culture
PC12 cells (Cell Resource Center, IBMS, CAMS/PUMC) were routinely cultured in RPMI-1640 medium (Gibco, Beijing, China) supplemented with 10% heat-inactivated fetal bovine serum (Gibco, Auckland, New Zealand), 5% heat-inactivated horse serum (Gibco, Auckland, New Zealand), 100 U/mL penicillin, and 100 μg/mL streptomycin (HyClone, South Logan, UT, USA) at 37 °C in a humidified atmosphere of 95% air and 5% CO2. PC12 cells (1×105 cells/mL) in low-serum RPMI-1640 medium (2% fetal bovine serum). Cells were seeded in 6-well plates or 96-well plates. The cells were allowed to grow for 24 h before processing for further experiments.
Drug treatment
FA (99.6% purity) was purchased from the Chinese National Institute for the Control of Pharmaceutical and Biological Products (Beijing, China). FA and rolipram were dissolved in dimethyl sulfoxide (DMSO) at 10-2 mol/L and then diluted in culture medium. The final concentration of DMSO used in the FA and rolipram treatment conditions was less than 0.1% LPS (Sigma, USA) was dissolved in sterile, pyrogen-free water and diluted with sterilized phosphate-buffered saline.
Treatment conditions
For PDE4B activity assays and RNA extraction, PC12 cells were pretreated with various concentrations of FA (0, 2.5, 5.0, 10, 20, or 40 μmol/L) or rolipram (30 μmol/L) for 12 h after stimulation with LPS (1 μg/mL) for 3 h. For Western blot and F-actin confocal analysis, cells were pretreated with various concentrations of FA or rolipram for 12 h and then incubated with LPS (1 μg/mL) for an additional 8 h35.
Immunofluorescence assay
Expression of F-actin was measured in PC12 cells which were pretreated with various concentrations of FA (10, 20, or 40 μmol/L) or rolipram (40 μmol/L) for 12 h and then stimulated with LPS for 8 h. The vehicle group (0.1% DMSO) and the FA (40 μmol/L) treatment group were not stimulated with LPS. The cultured PC12 cells were fixed with 4% paraformaldehyde in 0.1 mol/L phosphate buffer (pH 7.4; Invitrogen) for 15 min. After being washed 3 times with 0.1 mol/L phosphate buffer, blocked with 0.5% BSA in 0.1 mol/L phosphate buffer, and then permeabilized with 0.1% Triton X-100 (Solarbio, China) for 10 min at room temperature, cells were washed 3 times with 0.1 mol/L phosphate buffer and then incubated with 1 μmol/L Rhodamine phalloidin (Invitrogen) for 30 min at room temperature. Cells were then washed 3 times with 0.1 mol/L phosphate buffer, treated with DAPI (Solarbio, China) to stain nuclei, and washed another 3 times with 0.1 mol/L phosphate buffer. Cells were then examined with a laser-scanning confocal microscope with an excitation wavelength of 561 nm and an emission filter at 615±35 nm for rhodamine and an excitation filter at 405 nm and emission filter at 445±30 nm for DAPI (PerkinElmer UltraVIEW VoX Confocal Imaging System).
Cytokine assays
To further explore whether the neuroprotective effect of FA partly results from its anti-inflammatory effect, the culture medium from the PC12 cells was collected for pro-inflammatory cytokine assays. PC12 cells were pretreated with FA (2.5, 5, 10, 20, or 40 μmol/L) for 12 h, and this was followed by LPS (1 μg/mL) stimulation for 8 h. The control groups were a vehicle group (0.1% DMSO) and a group treated with FA (40 μmol/L) without LPS stimulation. The TNF-α and IL-1β levels in the cultured supernatant were quantified using an ELISA kit (Multiscience, China), per the manufacturer's instructions. The absorbance at 450 nm was determined using a VICTOR™ X5 Multilabel Plate Reader.
Phosphodiesterase activity assay
PC12 cells were grown in 60 mm dishes and pretreated with various concentrations of FA (0, 2.5, 5, 10, 20, or 40 μmol/L) and rolipram (30 μmol/L) for 12 h and then treated with LPS for 3 h. To examine whether FA could directly inhibit the cAMP-PDE activity, PC12 cells were treated with various concentrations of FA (0, 2.5, 5, 10, 20, or 40 μmol/L) alone for 12 h. PC12 cell cultures were lysed by the addition of cell lysis buffer (Applygen Technologies Inc, Beijing, China) containing a cocktail of protease inhibitors (Applygen, Beijing, China), and the total protein concentration was estimated using a BCA Protein Assay Kit (CW Biotech, Beijing, China). PDE4-specific enzymatic activity was determined using a PDE4 assay kit (PDE-Glo™, Promega Corporation). Each assay used 25 μg protein, which was diluted in 1× PDE-Glo™ Reaction Buffer; each assay was carried out in triplicate, per the manufacturer's instructions with some modifications, and luminescence was measured with a plate-reading luminometer.
RNA extraction and real-time RT-PCR
Total RNA was extracted from PC12 cells using TRIzol reagent (Sigma) for real-time PCR (Q-PCR), and the first-strand cDNA was reverse-transcribed into cDNA by using TransScrip First-Strand cDNA Synthesis SuperMix (TranGen Biotech, Beijing, China). The reverse transcription was performed using 10 ng of total RNA. The reverse transcription conditions were 30 min at 42 °C and 5 min at 85 °C. Gene transcripts were quantified by semi-quantitative real-time RT-PCR, which was performed in triplicate with an Applied Biosystems StepOnePlus Real-Time PCR System, with TransStart Green Q-PCR SuperMix (TranGen Biotech, Beijing, China) in 20 mL reaction volumes. Rat sequence-specific primers were synthesized by Invitrogen (Shanghai, China) as follows:
PDE4B-pan-specific forward, 5′-GACCGGATACAGGTTCTTCGA-3′, reverse, 5′-GAGTTCCCGGTTCAGCATCC-3′ β-actin forward, 5′-GTCGTACCACTGGCATTGTG-3′, reverse, 5′- GCTGTGGTGGTGAAGCTGTA-3′.
The fold changes in expression were determined with the comparative cycle threshold (CT) method (2-ΔΔCT), with normalization to β-actin as an endogenous reference gene in all the experiments. The results are presented as fold change over control.
Western blot analysis
Immunoblotting was used to quantify the protein levels of PDE4B, CREB and pCREB in LPS-activated PC12 cells. CREB and p-CREB are expressed in the nucleus, whereas PDE4B is a cytoplasmic protein. A Nuclear and Cytoplasmic Protein Extraction Kit (CW Biotech, Beijing, China) was used according to the manufacturer's instructions, and a protease inhibitor cocktail and phosphatase inhibitors were added. The supernatant (cytoplasmic extract) and the fraction containing nuclear proteins were stored in small aliquots at -80 °C until further use. The protein concentrations in extracts were measured using a BCA Protein Assay Kit (CW Biotech, Beijing, China), per the manufacturer's protocol.
Next, immunoblotting was performed according to the antibody manufacturers' instructions, using anti-CREB (48H2) rabbit mAb, anti-phospho-CREB (Ser133) (87G3) rabbit mAb (Cell Signaling Technology, Inc, Danvers, MA, USA), anti-PDE4B (Santa Cruz Biotechnology, Dallas, TX, USA), anti-β-actin (Cell Signaling Technology, Inc, Danvers, MA, USA) and anti-Histone H3 Rat Monoclonal Antibody (Bioeasy, Beijing, China). The bound primary antibodies were detected using horseradish peroxidase-conjugated anti-rabbit secondary antibody. Immunoreactive bands were visualized using Chemiluminescent HRP Substrate detection reagents (Millipore Corporation, Billerica, USA) and visualized with a ChemiScope3600 Mini chemiluminescence imaging system (Clinx Science Instruments, Shanghai, China). Histone H3 served as a loading control. Quantification was performed using ImageJ analysis software.
Docking
A crystal structure of PDE4B2 (PDB ID: 1R06) was downloaded from the protein data bank (PDB, http://www.rcsb.org/pdb/explore/explore.do?structureId=1RO6), which was used for molecular modeling studies36. The file of FA was obtained from the PubChem Compound Database (http://pubchem.ncbi.nlm.nih.gov/compound/445858#section=Top).
All the formats were saved in PDBQT format with AutoDock Tools (ADT). Molecular docking was performed with AutoDock 4.0 software (Scripps Research Institute, La Jolla, CA, USA) to generate an ensemble of docking conformations between FA and PDE4B2.
Statistical analysis
All data are expressed as the mean±SD, and at least three replicates were analyzed. Significant differences between experimental groups were determined using one-way ANOVA followed by Dunnett's test using GraphPad Prism 5 (GraphPad Software Inc, La Jolla, CA, USA). P values less than 0.05 were considered to be significant.
Results
Effect of FA on F-actin levels in LPS-stimulated PC12 cells
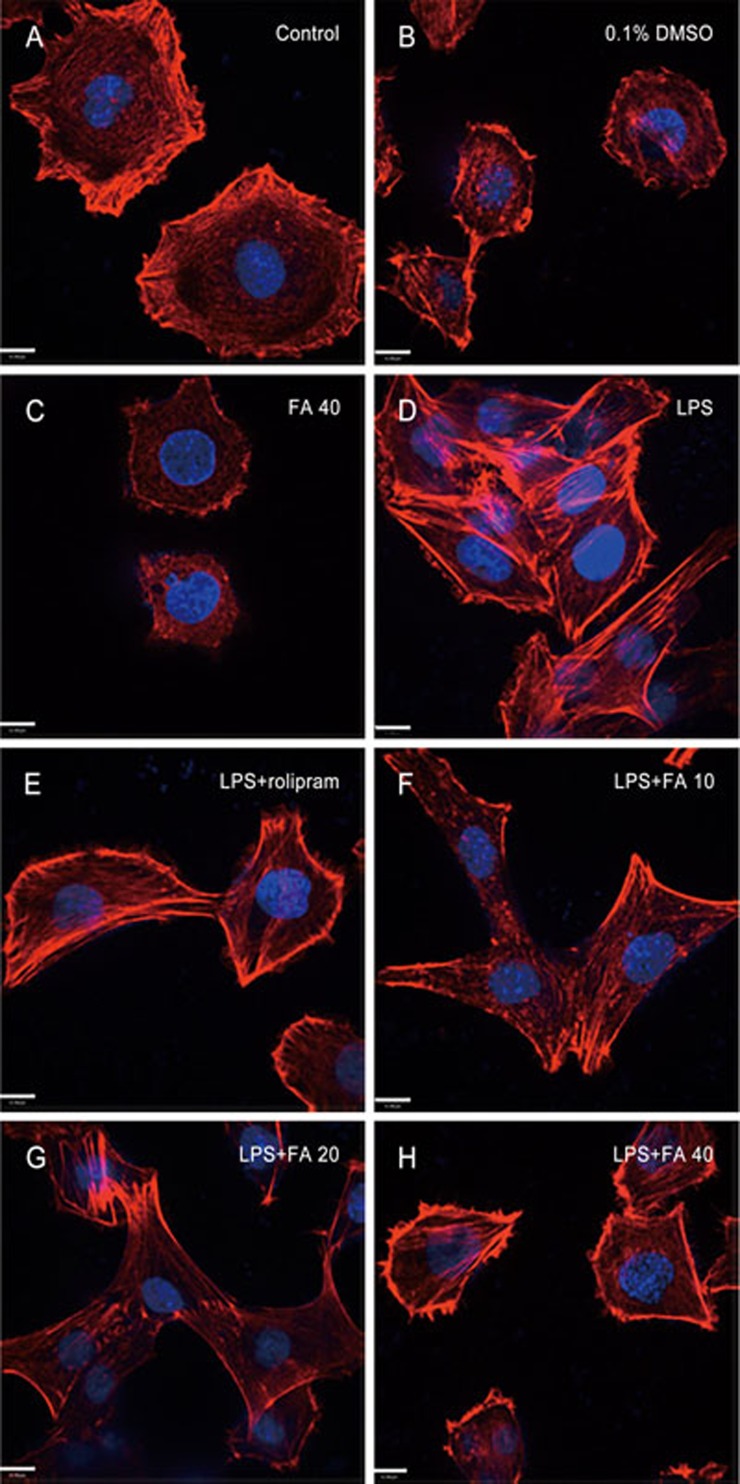
We examined the fluorescence intensity of F-actin. In PC12 cells treated with LPS (1 μg/mL) for 8 h, F-actin reorganized, and we observed a loss of fine F-actin stress fibers from the actin cytoskeleton (Figure 1D). Pretreatment with rolipram (30 μmol/L) and different concentrations of FA (10, 20, 40 μmol/L) for 12 h prevented the LPS-induced loss of fine F-actin stress fibers and appearance of thick bundles of actin filaments in PC12 cells, indicative of a healthy and well-developed actin cytoskeleton (Figure 1E–1H). F-actin in the vehicle group (0.1% DMSO) and the FA (40 μmol/L) group without LPS stimulation showed no obvious differences compared with the control group (Figure 1A, 1B, 1C).
Figure 1.
Effect of ferulic acid (FA) on F-actin level. Fluorescence staining of F-actin with Alexa Fluor 488 rhodamine-conjugated phalloidin (red) and of cell nuclei with DAPI (blue). (A–C) F-actin expression of PC12 cells in the control group, the vehicle group (0.1% DMSO) and the FA (40 μmol/L) group. (D) Changes in F-actin in PC12 cells induced by LPS (1 μg/mL) for 8 h. (E–H) PC12 cells were pretreated with rolipram (30 μmol/L) or FA (10, 20 or 40 μmol/L) for 12 h, and this was followed by LPS (0.15 mg/mL) treatment for 8 h. Scale bar, 11 μm (original magnification: ×630).
Pretreatment with FA inhibits the generation of TNF-α and IL-1β
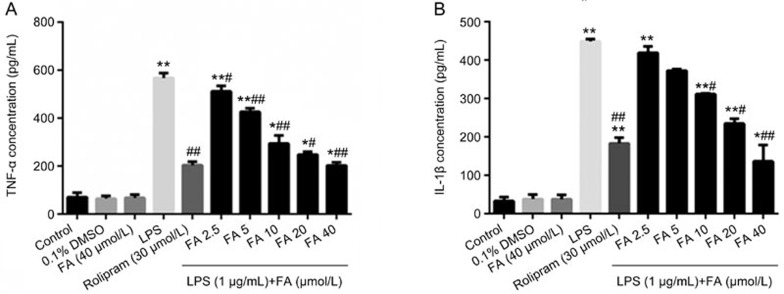
As shown in Figure 2A and 2B, LPS markedly increased the release of the pro-inflammatory cytokines TNF-α and IL-1β into the culture medium of PC12 cells compared with the control group. FA, compared with the LPS-treated group, significantly attenuated the release of TNF-α and IL-1β into the medium, and treatment with FA (40 μmol/L) alone or 0.1% DMSO without LPS stimulation did not significantly affect the release of these inflammatory mediators, as compared with the control group.
Figure 2.
Ferulic acid (FA) inhibits the generation of TNF-α and IL-1β. PC12 cells were pre-incubated for 12 h with the indicated concentrations of FA and then exposed to LPS (1 μg/mL) in culture for 8 h. The levels of TNF-α (A) and IL-1β (B) in the culture supernatants were determined. LPS significantly increased the TNF-α and IL-1β levels compared with those in the control group. Pretreatment with FA significantly decreased the TNF-α and IL-1β levels compared with those in the LPS group. Results are shown as the mean±SD of three experiments. *P<0.05, **P<0.01 vs control group. #P<0.05, ##P<0.01 vs LPS group.
Pretreatment with FA down-regulated cAMP-dependent PDE activity in LPS-stimulated PC12 cells
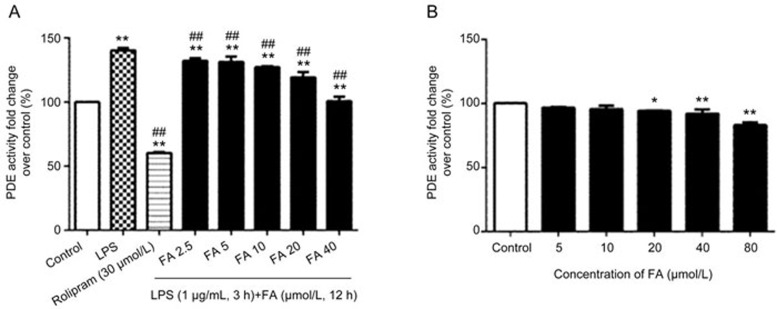
LPS treatment of PC12 cells up-regulated the cAMP-dependent PDE activity, which is critical for the PDE/cAMP/CREB signaling pathway. Therefore, we examined whether FA decreases the PDE4 activity induced by LPS. We measured the total cAMP-dependent PDE activity in lysates from PC12 cells that had been pretreated with various concentrations of FA for 12 h and then stimulated with LPS (1 μg/mL) for 3 h. As expected, LPS exposure led to the induction of PDE4 activity, resulting in a rapid (within 3 h), significant increase in cAMP-PDE activity as compared with that in the controls. FA pretreatment markedly attenuated the LPS-induced up-regulation of cAMP-PDE activity in PC12 cells (Figure 3A). Hence, we examined whether FA inhibits the cAMP-PDE activity directly. PC12 cells were treated with various concentrations of FA alone for 12 h, but no significant changes in cAMP-PDE activity were observed (Figure 3B).
Figure 3.
Effect of ferulic acid (FA) on LPS-induced cyclic AMP-dependent PDE activity. (A) LPS acutely increased cyclic AMP-dependent PDE activity in PC12 cells after 3 h of stimulation, but total cyclic AMP-dependent PDE activity decreased in PC12 cells pretreated with FA for 12 h. (B) Effect of FA on cyclic AMP-dependent PDE activity. PDE4 activity in PC12 cells treated with various concentrations of FA alone for 12 h. No significant decrease in cyclic AMP-dependent PDE activity was observed. Cyclic AMP-dependent PDE activity is expressed as the fold change over controls. Results are shown as the mean±SD of three experiments. *P<0.05, **P<0.01 vs control group. ##P<0.01 vs LPS group.
Pretreatment with FA inhibits the LPS-induced mRNA expression of PDE4B in PC12 cells
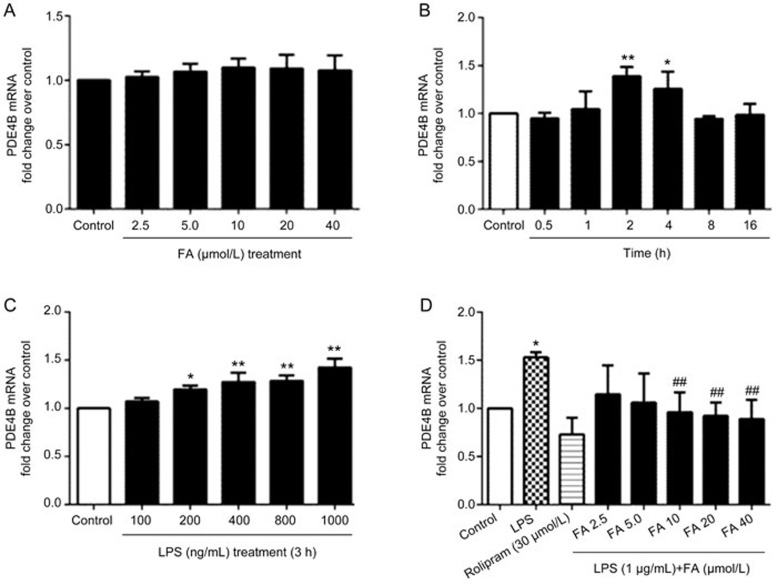
Changes in PDE4B mRNA levels were investigated using generic primers. The expression of PDE4B mRNA was measured at different time points from 30 min to 16 h after LPS treatment in PC12 cells. Expression of PDE4B mRNA was increased by LPS stimulation, but no significant increases in PDE4B mRNA were observed within 30 min of LPS stimulation at a dose of 1 μg/mL of LPS. The largest increase in PDE4B mRNA was observed at approximately 3 h after LPS treatment (Figure 4B and 4C). To determine whether the LPS-induced increase in PDE4B mRNA levels would be altered by FA, PC12 cells were pretreated with FA for 12 h and then stimulated with LPS for 3 h. PDE4B mRNA levels were altered by various concentrations of FA in the pretreatment (Figure 4D), but treatment with various concentrations of FA alone did not change the PDE4B mRNA expression (Figure 4A).
Figure 4.
Effect of ferulic acid (FA) on LPS-induced up-regulation of PDE4B mRNA expression. (A) PDE4B mRNA expression in PC12 cells after treatment with various concentrations of FA alone for 12 h. No significant changes in the PDE4B mRNA expression levels were observed. (B) PC12 cells were stimulated with LPS (1 μg/mL) for different lengths of time (0.5, 1, 2, 4, 8, or 16 h) before RNA extraction. No obvious increases in PDE4B mRNA expression were observed within 30 min of LPS stimulation. A significant increase in PDE4B mRNA expression was observed after approximately 3 h of treatment. (C) PC12 cells were stimulated with LPS (100 ng/mL–1 μg/mL) for 3 h. Expression of PDE4B was significantly increased by LPS at approximately 1 μg/mL. (D) PC12 cells showed significant decreases in PDE4B mRNA after pretreatment with various concentrations of FA for 12 h and stimulation with 1 μg/mL LPS for 3 h. Results are shown as the mean±SD of three experiments. *P<0.05, **P<0.01 vs control group. ##P<0.01 vs LPS group.
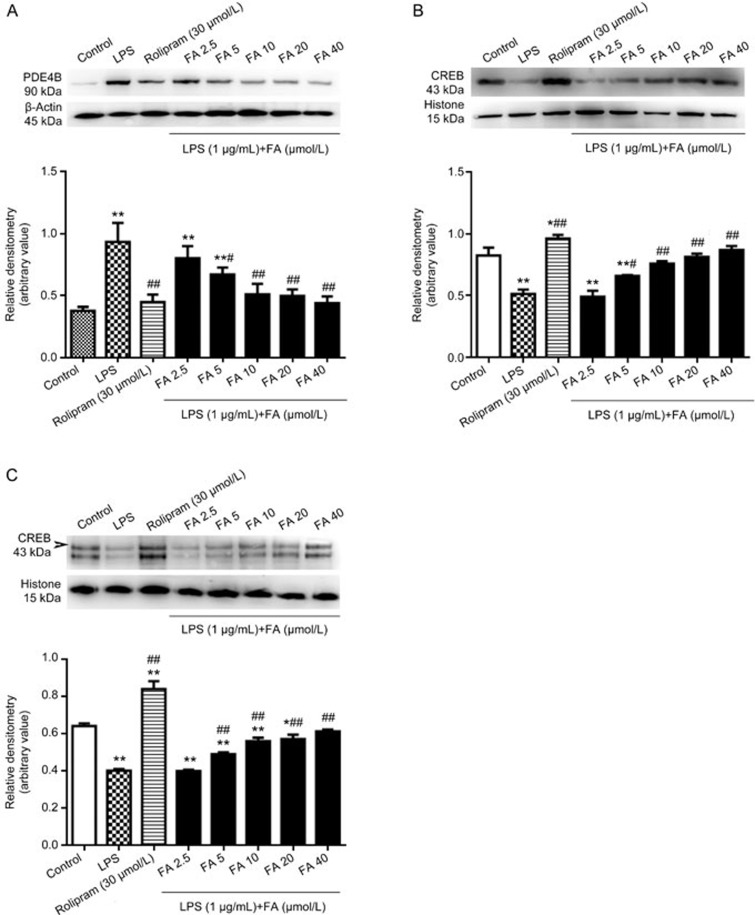
Effects of FA on the protein expression of PDE4B and CREB and the phosphorylation of CREB in LPS-stimulated PC12 cells
Western blot analysis showed that after LPS stimulation for 8 h in PC12 cells, PDE4B exhibited a significant increase in molecular weight, by approximately 90 kDa (Figure 5A). Pretreatment of PC12 cells with FA for 12 h decreased the LPS-induced PDE4B expression. CREB is a major downstream signaling molecule in the PDE4B/cAMP/CREB signaling pathway. To evaluate the activation of FA on the PDE4B/cAMP/CREB signaling cascade in PC12 cells, the levels of CREB and pCREB were detected by Western blotting, which showed that FA reversed the LPS-induced attenuation of CREB and pCREB expression. As shown in Figure 5B and 5C, 8 h of LPS exposure induced significant decreases in CREB and pCREB levels compared with those in the control group, but FA pretreatment increased the expression of CREB and pCREB compared with that in the LPS group.
Figure 5.
Effects of ferulic acid (FA) on the LPS-induced decreases in protein levels of PDE4B, CREB and phosphorylated CREB (pCREB). (A) PC12 cells were treated with 1 μg/mL LPS for 8 h after pretreatment with various concentrations of FA for 12 h. PDE4B expression levels decreased significantly. (B and C) Pretreatment with FA increased the protein levels of CREB and pCREB attenuated by LPS treatment. Results are shown as the mean±SD of three experiments. *P<0.05, **P<0.01 vs control group. #P<0.05, ##P<0.01 vs LPS group.
Molecular docking
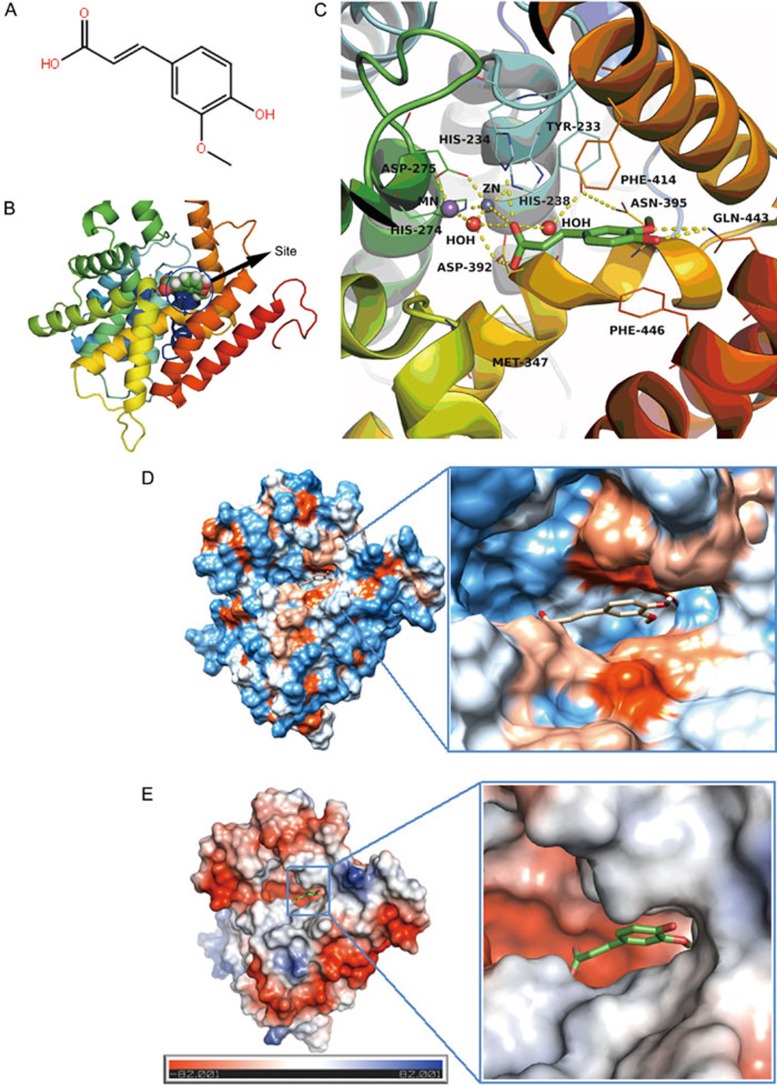
The molecular docking results are shown in Figure 6. This assay showed that FA interacts strongly with amino acid residues including Tyr233, His234, Met347, Asn395, Phe414, Gln443, and Phe446 at the FA-binding site of PDE4B2 and that FA can enter the binding cavity of PDE4B2 and form π-π interactions with the amino acid residues Phe446 and Phe414. Hydrogen bonding may be found between FA and amino acid residues Gln443 and His234 (Figure 6C). The area that interacts with FA is located in the hydrophobic cavity area, thus indicating that there are hydrophobic and electrostatic interactions between FA and PDE4B2 (Figure 6D and 6E), and these interactions contribute to the free energy of binding between FA and PDE4B2. The estimated free energy of binding (FEB) predicts the Van der Waals energy, electrostatic energy, and hydrogen bond energy. The most stable conformation exhibited the lowest FEB. The mean binding energy of FA with PDE4B2 is -6.36 kcal/mol, while the lowest bingding energy between them is -6.53 kcal/mol.
Figure 6.
Molecular docking results. (A) Chemical structure of ferulic acid (FA). (B) Predicted binding sites in PDE4B2. (C) Close view of binding mode of FA with PDE4B2 active site residues. Hydrogen bonds are represented by yellow dotted lines. (D) Hydrophobic interaction between FA and PDE4B2. (E) Electrostatic interaction between FA and PDE4B2.
Discussion
In living cells, the F-actin cytoskeleton encompasses multiple structures that are essential for different aspects of cell physiology. F-actin networks are involved in cell migration, cell protrusion, adhesion (lamellae and stress fibers), morphological changes (cortical actin) and cell division37. The cytoskeleton showed a dynamic change after LPS treatment of PC12 cells for 8 h: F-actin reorganized, and thick bundles of actin filaments appeared (Figure 1D). However, pretreatment of PC12 cells with rolipram and various concentrations of FA for 12 h prevented the LPS-induced loss of fine F-actin stress fibers and appearance of thick bundles of actin filaments, a result indicative of normal and well-developed F-actin (Figure 1E-1H).
Furthermore, we observed that pretreatment with FA significantly decreased the LPS-induced release of the pro-inflammatory cytokines TNF-α and IL-1β into the culture medium of PC12 cells. Cytokines play important roles in the organization and regulation of inflammatory responses38. Neuroinflammation is an important pathoetiologic hallmark of AD39,40. FA has been shown to ameliorate neuroinflammation and to decrease the expression of proinflammatory cytokines (TNF-α and IL-1β) in PSAPP mice41. Proinflammatory cytokines such as tumor necrosis factor (TNF)-α and interleukin (IL)-1β activate Toll-like receptor 4 (TLR4), thus culminating in the activation of nuclear factor (NF)-κB.
Studies have shown that ferulic acid inhibits the expression of the cytokines TNF-α and IL-1β by inhibiting their transcription42. Huang et al have investigated the inhibitory effect of FA on neuroinflammation in BV-2 microglial cells induced by LPS. They have shown that that FA significantly suppresses the production of nitric oxide (NO), prostaglandin E2 (PGE2), and IL-1β and decreases the induction of type II nitric oxide synthase (iNOS) and cyclooxygenase-2 (COX-2) protein expression in LPS-stimulated BV-2 microglia in a dose-dependent manner. They hypothesized that this effect is achieved by suppression of the protein expression of TLR443.
As shown in this study, compared with LPS alone, pretreated with FA or rolipram inhibited the production of PDE4B, but the inhibitory effect of FA in PC12 cells was considerably milder than that of rolipram. However, FA increased the production of CREB and pCREB in response to LPS stimulation at levels almost comparable to those induced by rolipram. These data suggested that the inhibitory effects of FA might be different from those of rolipram, such that FA more selectively affects certain steps in the LPS-induced production of inflammatory factors, in contrast to rolipram, which inhibits the activity of PDE4B by binding to it tightly. Evidence has shown that TNF-α and IL-β are potent cytokines that induce of PDE4B as well as cAMP44. In this study, we confirmed that LPS at 1 μg/mL is sufficient to induce an increase in mRNA expression of PDE4B in PC12 cells. The analyses of PDE4B mRNA expression over time indicated that PDE4B mRNA levels increased significantly at approximately 3 h after stimulation with LPS, whereas PDE4B mRNA appeared at levels similar to baseline within the first half hour and after 8 h of treatment with LPS in PC12 cells.
PDE4 is the major cAMP-hydrolyzing enzyme found in inflammatory and immune cells45,46. The PDE4 family is composed of four isozymes, PDE4A to D47. These isozymes are widely expressed in many tissues, and high concentrations found in the brain48,49. PDE4 isozymes are involved in different functions, depending on their patterns of distribution in the brain50,51. The PDE4 subtypes are distributed in different inflammatory cells, and they appear to be important regulators of inflammatory processes; PDE4 is involved in many disorders such as depression, anxiety, and inflammation-related disorders51,52. Selective inhibitors of PDE4 have been suggested for the treatment of a variety of CNS diseases, including Alzheimer's disease, depression, Parkinson's disease and Huntington's disease53,54,55.
In our present investigation, we hypothesized that FA could inhibit LPS-induced PDE4B expression (Figure 5A), even though treatment with FA alone might have only a weak influence on the enzyme activity of PDE4B in PC12 cells (Figure 3B). An inhibitory effect of FA on PDE4B production was shown in this study, and the present results suggest that PDE4B production might be induced directly by LPS but also partly indirectly via the initial induction of TNF-α and IL-1β. It has been reported that IL-1β production is induced by LPS itself and that the second phase of the inflammatory response is due to the LPS-stimulated release of mediators such as TNF-α and IL-1β44. In PSAPP mice, FA ameliorates neuro-inflammation and decreases the expression of pro-inflammatory cytokines (TNF-α and IL-1β)56. Furthermore, we observed that pretreatment with FA significantly and dose-dependently decreased the LPS- and Aβ25-35-induced release of the pro-inflammatory cytokines TNF-α and IL-1β into the culture medium of PC12 cells33.
We further examined the effects of FA on the production of CREB and pCREB in addition to the expression of PDE4B in LPS-stimulated PC12 cells. The results demonstrated that the expression levels of CREB and pCREB were maintained by FA pretreatment in LPS-stimulated PC12 cells. The PDE4B/cAMP/CREB pathway plays an important role in modulating various inflammatory reactions; thus, the inhibition of cytokine production is expected to exhibit an anti-inflammatory effect through the reduction of PDE4B expression, thereby increasing the cAMP level at inflammatory sites. Our studies showed that pretreatment with FA inhibited PDE4B production and increased the production of both CREB and pCREB in LPS-stimulated PC12 cells.
CREB was first described by Montminy and Bilezikjian in 1987 as a cellular transcription factor that binds the cAMP-response element and leads to increased transcription of the somatostatin gene57. Evidence from studies in a wide range of species and using genetic and pharmacological manipulations of CREB has shown that CREB is essential for memory formation58,59,60,61. cAMP stimulates the transcription factor CREB by activating cAMP-dependent protein kinase, which regulates the transcription of many genes, including brain-derived neurotrophic factor (BDNF).
PDE4, which specifically hydrolyzes cAMP, regulates many crucial signaling cascades involved in learning and memory62,63,64. The ability of PDE4 inhibitors to enhance cognition appears to be attributed to the stimulation of the cAMP/CREB signaling pathway in the hippocampus. PDE4 inhibitors, such as rolipram, increase the cAMP concentration and the phosphorylation of CREB, which acts as a transcriptional activator only after it is phosphorylated by certain protein kinases. Increasing the CREB levels enhances some forms of long-term memory and improves spatial and associative memory in mice65,66,67,68. In this study, the levels of CREB and pCREB in LPS-stimulated PC12 cells were much lower than those in the FA-pretreated group. These data suggest that the contribution of FA to the PDE4B/cAMP/CREB pathway reduces the activity of PDE4B in this experimental system.
Molecular docking results indicated the presence of hydrogen bonds, π-π conjugate electrostatic and hydrophobic interactions between FA and PDE4B2. Although further studies should be carried out to confirm these interactions, this result of molecular docking provides theoretical support for the potential effects of FA on PDE4B enzymatic activity and expression. These results indicated that FA may be used as a basic structure for designing PDE4B inhibitors.
The results of this study provide the first demonstration that one of the anti-inflammatory mechanisms of FA can be attributed to its ability to inhibit LPS-induced PDE4B activity and expression. PDE4B appears to be the major PDE4 enzyme subtype responsible for cAMP hydrolysis because much evidence has demonstrated that the inhibition or ablation of PDE4B produces a broad spectrum of anti-inflammatory effects while minimizing unwanted side effects67,69. The development of PDE4 inhibitors with PDE4B selectivity has been considered to be a promising approach for treatment of neuro-inflammatory diseases such as AD, for which the PDE4B isozyme family may be a useful therapeutic target.
FA is an enhancer of cAMP, and cAMP enhancers have been found to suppress the production of TNF stimulated either in vitro or in vivo with LPS70. Various studies have documented that suppression of LPS-inducible TNF production by cAMP can occur via both NF-κB-dependent and -independent mechanisms71. To clarify the neuroprotective mechanism of FA, further studies are needed to explain the interaction between the LPS pathway and the cAMP pathway.
In summary, the present findings showed that FA effectively inhibits LPS-induced PDE4B expression, along with its effect of increasing CREB and pCREB, FA may be an attractive therapeutic agent for treatment of memory and cognition impairment associated with age-dependent or neurodegenerative diseases. However, the mechanisms of the anti-inflammatory and neuro-protective effects of FA are not completely understood and require further study.
Author contribution
Hao HUANG, Qian HONG, and Yue GAO designed the research; Hao HUANG performed the experiments; Hong-ling TAN and Cheng-rong XIAO contributed some reagents and analytical tools; Hao HUANG analyzed the data; and Hao HUANG wrote the paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No 81130067 and 81202936).
References
- Eikelenboom P, Veerhuis R, Scheper W, Rozemuller AJ, van Gool WA, Hoozemans JJ. The significance of neuroinflammation in understanding Alzheimer's disease. J Neural Transm 2006; 113: 1685–95. [DOI] [PubMed] [Google Scholar]
- Sayyah M, Javad-Pour M, Ghazi-Khansari M. The bacterial endotoxin lipopolysaccharide enhances seizure susceptibility in mice: involvement of proinflammatory factors: nitric oxide and prostaglandins. Neuroscience 2003; 122: 1073–80. [DOI] [PubMed] [Google Scholar]
- Jaeger LB, Dohgu S, Sultana R, Lynch JL, Owen JB, Erickson MA, et al. Lipopolysaccharide alters the blood-brain barrier transport of amyloid beta protein: a mechanism for inflammation in the progression of Alzheimer's disease. Brain Behav Immun 2009; 23: 507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi N, Guroff G. Nerve growth factor-induced increase in the cell-free phosphorylation of a nuclear protein in PC12 cells. J Biol Chem 1985; 260: 7791–9. [PubMed] [Google Scholar]
- Isoda H, Talorete TP, Kimura M, Maekawa T, Inamori Y, Nakajima N, et al. Phytoestrogens genistein and daidzin enhance the acetylcholinesterase activity of the rat pheochromocytoma cell line PC12 by binding to the estrogen receptor. Cytotechnology 2002; 40: 117–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Li F, Wu Q, Gong Q, Lu Y, Shi J. Protective effects of icariin on brain dysfunction induced by lipopolysaccharide in rats. Phytomedicine 2010; 17: 950–5. [DOI] [PubMed] [Google Scholar]
- Min SS, Quan HY, Ma J, Han JS, Jeon BH, Seol GH. Chronic brain inflammation impairs two forms of long-term potentiation in the rat hippocampal CA1 area. Neurosci Lett 2009; 456: 20–4. [DOI] [PubMed] [Google Scholar]
- Breder CD, Saper CB. Expression of inducible cyclooxygenase mRNA in the mouse brain after systemic administration of bacterial lipopolysaccharide. Brain Res 1996; 713: 64–9. [DOI] [PubMed] [Google Scholar]
- Breder CD, Hazuka C, Ghayur T, Klug C, Huginin M, Yasuda K, et al. Regional induction of tumor necrosis factor alpha expression in the mouse brain after systemic lipopolysaccharide administration. Proc Natl Acad Sci U S A 1994; 91: 11393–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Irisarri E, Perez-Torres S, Miro X, Martinez E, Puigdomenech P, Palacios JM, et al. Differential distribution of PDE4B splice variant mRNAs in rat brain and the effects of systemic administration of LPS in their expression. Synapse 2008; 62: 74–9. [DOI] [PubMed] [Google Scholar]
- Jin SL, Conti M. Induction of the cyclic nucleotide phosphodiesterase PDE4B is essential for LPS-activated TNF-alpha responses. Proc Natl Acad Sci U S A 2002; 99: 7628–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MM, Gristwood RW, Cooper N, Hellewell PG. Phosphodiesterase (PDE)4 inhibitors: anti-inflammatory drugs of the future? Trends Pharmacol Sci 1997; 18: 164–71. [DOI] [PubMed] [Google Scholar]
- Wang P, Wu P, Ohleth KM, Egan RW, Billah MM. Phosphodiesterase 4B2 is the predominant phosphodiesterase species and undergoes differential regulation of gene expression in human monocytes and neutrophils. Mol Pharmacol 1999; 56: 170–4. [DOI] [PubMed] [Google Scholar]
- Xu Y, Zhang HT, O'Donnell JM. Phosphodiesterases in the central nervous system: implications in mood and cognitive disorders. Handb Exp Pharmacol 2011; 204: 447–85. [DOI] [PubMed] [Google Scholar]
- Cherry JA, Davis RL. Cyclic AMP phosphodiesterases are localized in regions of the mouse brain associated with reinforcement, movement, and affect. J Comp Neurol 1999; 407: 287–301. [PubMed] [Google Scholar]
- Johansson EM, Reyes-Irisarri E, Mengod G. Comparison of cAMP-specific phosphodiesterase mRNAs distribution in mouse and rat brain. Neurosci Lett 2012; 525: 1–6. [DOI] [PubMed] [Google Scholar]
- Perez-Torres S, Miro X, Palacios JM, Cortes R, Puigdomenech P, Mengod G. Phosphodiesterase type 4 isozymes expression in human brain examined by in situ hybridization histochemistry and (3H] rolipram binding autoradiography. Comparison with monkey and rat brain. J Chem Neuroanat 2000; 20: 349–74. [DOI] [PubMed] [Google Scholar]
- Ahmed T, Frey JU. Expression of the specific type IV phosphodiesterase gene PDE4B3 during different phases of long-term potentiation in single hippocampal slices of rats in vitro. Neuroscience 2003; 117: 627–38. [DOI] [PubMed] [Google Scholar]
- Ahmed T, Frey JU. Phosphodiesterase 4B (PDE4B) and cAMP-level regulation within different tissue fractions of rat hippocampal slices during long-term potentiation in vitro. Brain Res 2005; 1041: 212–22. [DOI] [PubMed] [Google Scholar]
- Ahmed T, Frey S, Frey JU. Regulation of the phosphodiesterase PDE4B3-isotype during long-term potentiation in the area dentata in vivo. Neuroscience 2004; 124: 857–67. [DOI] [PubMed] [Google Scholar]
- Murakami A, Kadota M, Takahashi D, Taniguchi H, Nomura E, Hosoda A, et al. Suppressive effects of novel ferulic acid derivatives on cellular responses induced by phorbol ester, and by combined lipopolysaccharide and interferon-gamma. Cancer Lett 2000; 157: 77–85. [DOI] [PubMed] [Google Scholar]
- Hosoda A, Ozaki Y, Kashiwada A, Mutoh M, Wakabayashi K, Mizuno K, et al. Syntheses of ferulic acid derivatives and their suppressive effects on cyclooxygenase-2 promoter activity. Bioorg Med Chem 2002; 10: 1189–96. [DOI] [PubMed] [Google Scholar]
- Ou L, Kong LY, Zhang XM, Niwa M. Oxidation of ferulic acid by Momordica charantia peroxidase and related anti-inflammation activity changes. Biol Pharm Bull 2003; 26: 1511–6. [DOI] [PubMed] [Google Scholar]
- Srinivasan M, Sudheer AR, Menon VP. Ferulic acid: therapeutic potential through its antioxidant property. J Clin Biochem Nutr 2007; 40: 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TY, Lu CW, Huang SK, Wang SJ. Ferulic acid suppresses glutamate release through inhibition of voltage-dependent calcium entry in rat cerebrocortical nerve terminals. J Med Food 2013; 16: 112–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso C, Scapagini G, Curro D, Giuffrida Stella AM, De Marco C, Butterfield DA, et al. Mitochondrial dysfunction, free radical generation and cellular stress response in neurodegenerative disorders. Fronti Biosci 2007; 12: 1107–23. [DOI] [PubMed] [Google Scholar]
- Dong GC, Kuan CY, Subramaniam S, Zhao JY, Sivasubramaniam S, Chang HY, et al. A potent inhibition of oxidative stress induced gene expression in neural cells by sustained ferulic acid release from chitosan based hydrogel. Mater Sci EngC Materi Biol Appl 2015; 49: 691–9. [DOI] [PubMed] [Google Scholar]
- Cheng CY, Su SY, Tang NY, Ho TY, Chiang SY, Hsieh CL. Ferulic acid provides neuroprotection against oxidative stress-related apoptosis after cerebral ischemia/reperfusion injury by inhibiting ICAM-1 mRNA expression in rats. Brain Res 2008; 1209: 136–50. [DOI] [PubMed] [Google Scholar]
- Kawabata K, Yamamoto T, Hara A, Shimizu M, Yamada Y, Matsunaga K, et al. Modifying effects of ferulic acid on azoxymethane-induced colon carcinogenesis in F344 rats. Cancer Lett 2000; 157: 15–21. [DOI] [PubMed] [Google Scholar]
- Balasubashini MS, Rukkumani R, Viswanathan P, Menon VP. Ferulic acid alleviates lipid peroxidation in diabetic rats. Phytother Res 2004; 18: 310–4. [DOI] [PubMed] [Google Scholar]
- Sultana R, Ravagna A, Mohmmad-Abdul H, Calabrese V, Butterfield DA. Ferulic acid ethyl ester protects neurons against amyloid beta- peptide(1-42)-induced oxidative stress and neurotoxicity: relationship to antioxidant activity. J Neurochem 2005; 92: 749–58. [DOI] [PubMed] [Google Scholar]
- Yogeeta SK, Gnanapragasam A, Kumar SS, Subhashini R, Sathivel A, Devaki T. Synergistic interactions of ferulic acid with ascorbic acid: its cardioprotective role during isoproterenol induced myocardial infarction in rats. Mol Cell Biochem 2006; 283: 139–46. [DOI] [PubMed] [Google Scholar]
- Huang H, Ma ZC, Wang YG, Hong Q, Tan HL, Xiao CR, et al. Ferulic acid alleviates Abeta25-35- and lipopolysaccharide-induced PC12 cellular damage: a potential role in Alzheimer's disease by PDE inhibition. Int J Clin Pharmacol Ther 2015; 53: 828–37. [DOI] [PubMed] [Google Scholar]
- Yabe T, Hirahara H, Harada N, Ito N, Nagai T, Sanagi T, et al. Ferulic acid induces neural progenitor cell proliferation in vitro and in vivo. Neuroscience 2010; 165: 515–24. [DOI] [PubMed] [Google Scholar]
- Gobejishvili L, Avila DV, Barker DF, Ghare S, Henderson D, Brock GN, et al. S-adenosylmethionine decreases lipopolysaccharide-induced phosphodiesterase 4B2 and attenuates tumor necrosis factor expression via cAMP/protein kinase A pathway. J Pharmacol Exp Ther 2011; 337: 433–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RX, Rocque WJ, Lambert MH, Vanderwall DE, Luther MA, Nolte RT. Crystal structures of the catalytic domain of phosphodiesterase 4B complexed with AMP, 8-Br-AMP, and rolipram. J Mol Biol 2004; 337: 355–65. [DOI] [PubMed] [Google Scholar]
- Li S, Duance VC, Blain EJ. F-actin cytoskeletal organization in intervertebral disc health and disease. Biochem Soc Trans 2007; 35: 683–5. [DOI] [PubMed] [Google Scholar]
- Niemand C, Nimmesgern A, Haan S, Fischer P, Schaper F, Rossaint R, et al. Activation of STAT3 by IL-6 and IL-10 in primary human macrophages is differentially modulated by suppressor of cytokine signaling 3. J Immunol 2003; 170: 3263–72. [DOI] [PubMed] [Google Scholar]
- Akiyama H, Barger S, Barnum S, Bradt B, Bauer J, Cole GM, et al. Inflammation and Alzheimer's disease. Neurobio Aging 2000; 21: 383–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan V, Chauhan A. Oxidative stress in Alzheimer's disease. Pathophysiology 2006; 13: 195–208. [DOI] [PubMed] [Google Scholar]
- Cheng CY, Ho TY, Lee EJ, Su SY, Tang NY, Hsieh CL. Ferulic acid reduces cerebral infarct through its antioxidative and anti-inflammatory effects following transient focal cerebral ischemia in rats. Am J Chin Med 2008; 36: 1105–19. [DOI] [PubMed] [Google Scholar]
- Navarrete S, Alarcon M, Palomo I. Aqueous extract of tomato (Solanum lycopersicum L) and ferulic acid reduce the expression of TNF-alpha and IL-1beta in LPS-activated macrophages. Molecules 2015; 20: 15319–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Deng HM, Zhu MM, Xiao F, Yang L, Zhang ZJ, et al. Inhibitory effect of ferulic acid on inflammatory response in microglia induced by lipopolysaccharides. Dongwuxue Yanjiu 2011; 32: 311–6. [DOI] [PubMed] [Google Scholar]
- Ghosh M, Garcia-Castillo D, Aguirre V, Golshani R, Atkins CM, Bramlett HM, et al. Proinflammatory cytokine regulation of cyclic AMP-phosphodiesterase 4 signaling in microglia in vitro and following CNS injury. Glia 2012; 60: 1839–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumer W, Hoppmann J, Rundfeldt C, Kietzmann M. Highly selective phosphodiesterase 4 inhibitors for the treatment of allergic skin diseases and psoriasis. Inflamm Allergy Drug Targets 2007; 6: 17–26. [DOI] [PubMed] [Google Scholar]
- Souness JE, Aldous D, Sargent C. Immunosuppressive and anti-inflammatory effects of cyclic AMP phosphodiesterase (PDE) type 4 inhibitors. Immunopharmacology 2000; 47: 127–62. [DOI] [PubMed] [Google Scholar]
- Conti M, Richter W, Mehats C, Livera G, Park JY, Jin C. Cyclic AMP-specific PDE4 phosphodiesterases as critical components of cyclic AMP signaling. J Biol Chem 2003; 278: 5493–6. [DOI] [PubMed] [Google Scholar]
- D'Sa C, Eisch AJ, Bolger GB, Duman RS. Differential expression and regulation of the cAMP-selective phosphodiesterase type 4A splice variants in rat brain by chronic antidepressant administration. Eur J Neurosci 2005; 22: 1463–75. [DOI] [PubMed] [Google Scholar]
- McPhee I, Cochran S, Houslay MD. The novel long PDE4A10 cyclic AMP phosphodiesterase shows a pattern of expression within brain that is distinct from the long PDE4A5 and short PDE4A1 isoforms. Cell Signal 2001; 13: 911–8. [DOI] [PubMed] [Google Scholar]
- Miro X, Perez-Torres S, Puigdomenech P, Palacios JM, Mengod G. Differential distribution of PDE4D splice variant mRNAs in rat brain suggests association with specific pathways and presynaptical localization. Synapse 2002; 45: 259–69. [DOI] [PubMed] [Google Scholar]
- Zhang HT. Cyclic AMP-specific phosphodiesterase-4 as a target for the development of antidepressant drugs. Curr Pharm Des 2009; 15: 1688–98. [DOI] [PubMed] [Google Scholar]
- Houslay MD, Schafer P, Zhang KY. Keynote review: phosphodiesterase-4 as a therapeutic target. Drug Discov Today 2005; 10: 1503–19. [DOI] [PubMed] [Google Scholar]
- O'Donnell JM, Zhang HT. Antidepressant effects of inhibitors of cAMP phosphodiesterase (PDE4). Trends Pharmacol Sci 2004; 25: 158–63. [DOI] [PubMed] [Google Scholar]
- Titus SA, Li X, Southall N, Lu J, Inglese J, Brasch M, et al. A cell-based PDE4 assay in 1536-well plate format for high-throughput screening. J Biomol Screen 2008; 13: 609–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar JK, Mackie S, Clapcote SJ, Murdoch H, Pickard BS, Christie S, et al. Disrupted in schizophrenia 1 and phosphodiesterase 4B: towards an understanding of psychiatric illness. J Physiol 2007; 584: 401–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Koyama N, Guillot-Sestier MV, Tan J, Town T. Ferulic acid is a nutraceutical beta-secretase modulator that improves behavioral impairment and alzheimer-like pathology in transgenic mice. PLoS One 2013; 8: e55774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima T, Uchida C, Anderson SF, Parvin JD, Montminy M. Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev 1997; 11: 738–47. [DOI] [PubMed] [Google Scholar]
- Kida S, Josselyn SA, Pena de Ortiz S, Kogan JH, Chevere I, Masushige S, et al. CREB required for the stability of new and reactivated fear memories. Nat Neurosci 2002; 5: 348–55. [DOI] [PubMed] [Google Scholar]
- Josselyn SA, Kida S, Silva AJ. Inducible repression of CREB function disrupts amygdala-dependent memory. Neurobiol Learn Mem 2004; 82: 159–63. [DOI] [PubMed] [Google Scholar]
- Pittenger C, Huang YY, Paletzki RF, Bourtchouladze R, Scanlin H, Vronskaya S, et al. Reversible inhibition of CREB/ATF transcription factors in region CA1 of the dorsal hippocampus disrupts hippocampus-dependent spatial memory. Neuron 2002; 34: 447–62. [DOI] [PubMed] [Google Scholar]
- Restivo L, Tafi E, Ammassari-Teule M, Marie H. Viral-mediated expression of a constitutively active form of CREB in hippocampal neurons increases memory. Hippocampus 2009; 19: 228–34. [DOI] [PubMed] [Google Scholar]
- Ahmed T, Frey JU. Plasticity-specific phosphorylation of CaMKII, MAP-kinases and CREB during late-LTP in rat hippocampal slices in vitro. Neuropharmacology 2005; 49: 477–92. [DOI] [PubMed] [Google Scholar]
- Blokland A, Schreiber R, Prickaerts J. Improving memory: a role for phosphodiesterases. Curr Pharm Des 2006; 12: 2511–23. [DOI] [PubMed] [Google Scholar]
- Coskran TM, Morton D, Menniti FS, Adamowicz WO, Kleiman RJ, Ryan AM, et al. Immunohistochemical localization of phosphodiesterase 10A in multiple mammalian species. J Histochem Cytochem 2006; 54: 1205–13. [DOI] [PubMed] [Google Scholar]
- Han JH, Kushner SA, Yiu AP, Cole CJ, Matynia A, Brown RA, et al. Neuronal competition and selection during memory formation. Science 2007; 316: 457–60. [DOI] [PubMed] [Google Scholar]
- Jasnow AM, Shi C, Israel JE, Davis M, Huhman KL. Memory of social defeat is facilitated by cAMP response element-binding protein overexpression in the amygdala. Behav Neurosci 2005; 119: 1125–30. [DOI] [PubMed] [Google Scholar]
- Naganuma K, Omura A, Maekawara N, Saitoh M, Ohkawa N, Kubota T, et al. Discovery of selective PDE4B inhibitors. Bio Med Chem Lett 2009; 19: 3174–6. [DOI] [PubMed] [Google Scholar]
- Wallace TL, Stellitano KE, Neve RL, Duman RS. Effects of cyclic adenosine monophosphate response element binding protein overexpression in the basolateral amygdala on behavioral models of depression and anxiety. Biol Psychiatry 2004; 56: 151–60. [DOI] [PubMed] [Google Scholar]
- Maurice DH, Ke H, Ahmad F, Wang Y, Chung J, Manganiello VC. Advances in targeting cyclic nucleotide phosphodiesterases. Nat Rev Drug Discov 2014; 13: 290–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidek Z. Adenosine - cyclic AMP pathways and cytokine expression. Eur Cytokine Netw 1999; 10: 319–28. [PubMed] [Google Scholar]
- Gobejishvili L, Barve S, Joshi-Barve S, Uriarte S, Song Z, McClain C. Chronic ethanol-mediated decrease in cAMP primes macrophages to enhanced LPS-inducible NF-kappaB activity and TNF expression: relevance to alcoholic liver disease. Am J Physiol Gastrointest Liver Physiol 2006; 291: G681–8. [DOI] [PubMed] [Google Scholar]