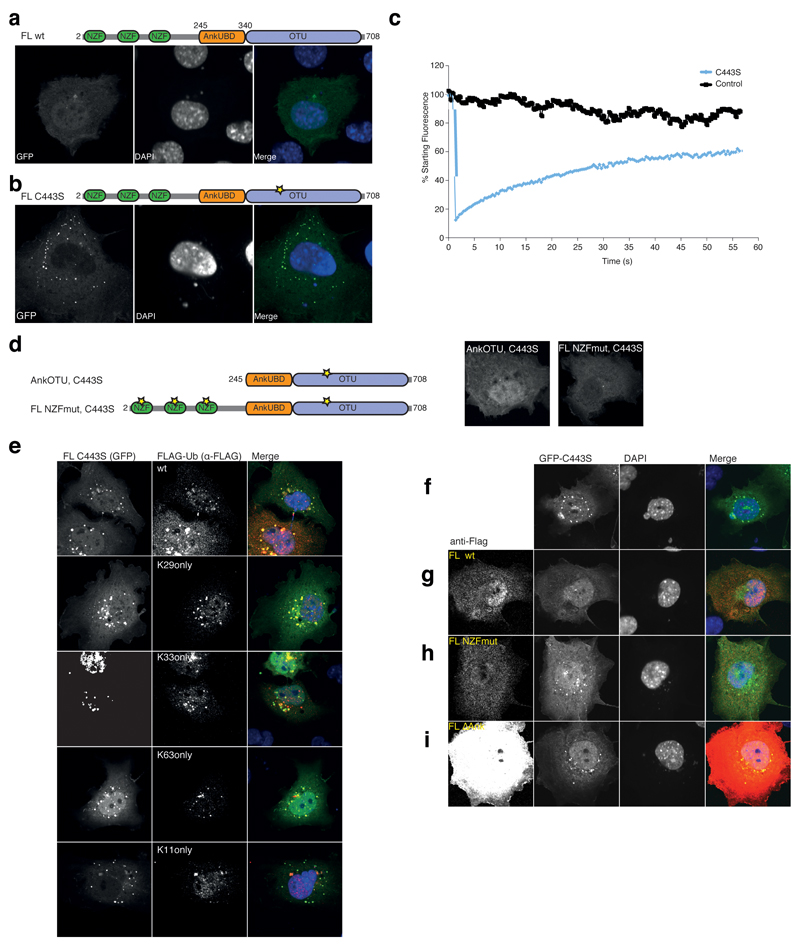
Figure 7.
In vivo DUB assay NZF and AnkUBD are essential for TRABID puncta. (a) Localization studies with GFP-TRABID in COS-7 cells 18 h after transfection (left). Nuclei are stained using DAPI (middle). The right image is a merge of the channels. The domain structure of TRABID is shown above. (b) A GFP-tagged full-length TRABID catalytic mutant (C443S, a yellow star in the domain representation indicates the mutation) adopts a punctate localization in COS-7 (shown) and other cell types 35. (c) Fluorescence recovery after photobleaching (FRAP) experiments on a control (black) or puncta containing volume (C443S, blue). Fluorescent recovery is recorded over time. (d) Localization studies of TRABID GFP-AnkOTU C443S and GFP-FL NZFmut C443S as in a. The domain structure is shown (left), and the GFP fluorescence of either construct (right) shows that no puncta are formed. (e) Puncta-forming GFP-C443S (left image) was co-expressed with FLAG-Ub or single-Lys (Konly) Ub mutants (middle image). The merged image is shown to the right. Further Ub mutants are shown in Supplementary Fig. 6a. (f-i) Dissolving TRABID assemblies requires the AnkUBD. (f) Puncta-forming GFP-tagged full-length TRABID C443S (left) was expressed in COS-7 cells and TRABID assemblies were visualized (left). Nuclei are stained using DAPI (middle). The merged image is shown to the right. (g-i) As in f, but in addition, FLAG-tagged TRABID constructs FL wt (g), FL NZFmut (h) or FL ∆Ank (i) were co-expressed (far left) and the presence of GFP puncta was assessed. Additional data is shown in Supplementary Fig. 6b.