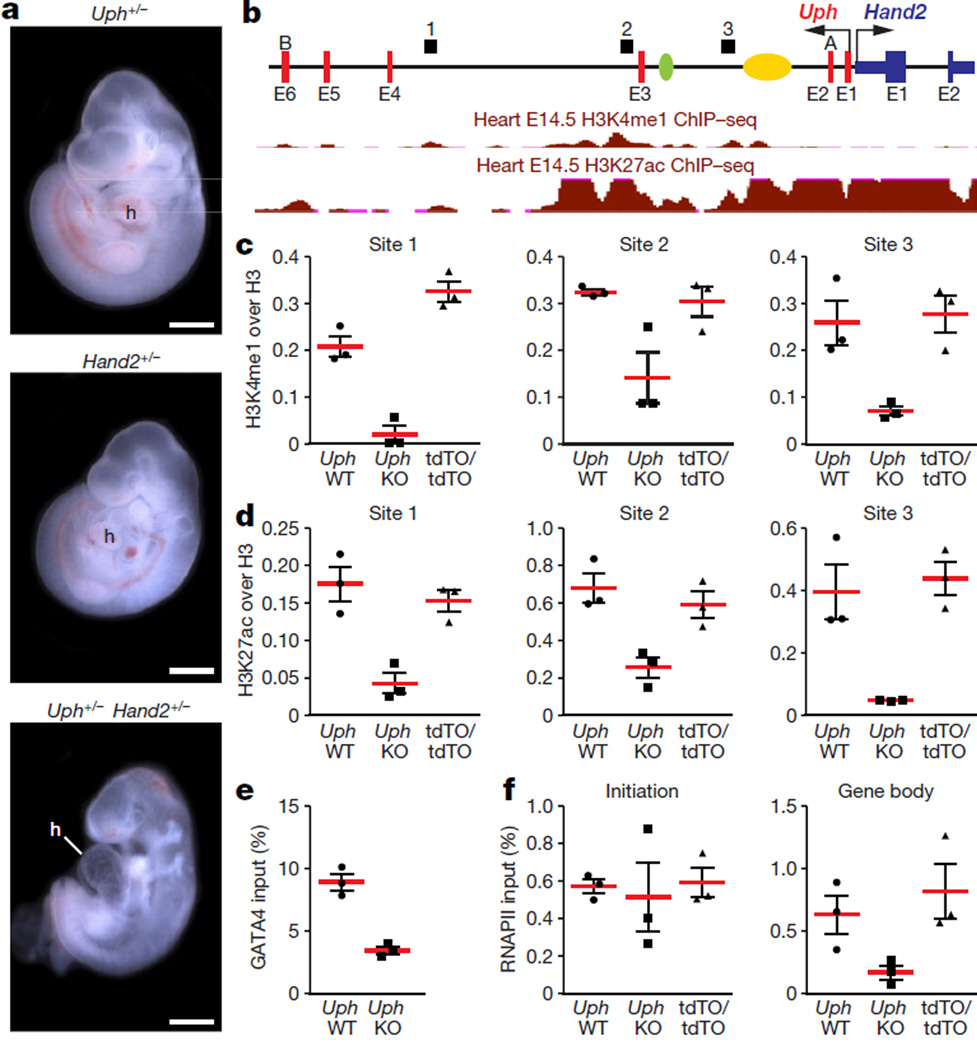
Figure 4. Uph transcription in cis is required for active enhancer marks in the Uph locus and RNAPII elongation at the Hand2 gene.
a, E10.5 embryos heterozygous for either Uph (top) or Hand2 (middle) appear normal in size and morphology. By contrast, Uph+/− Hand2+/− double heterozygote embryos developed a single ventricle. Scale bars, 1 mm. b, Uph–Hand2 locus with letters (A or B) indicating GapmeR target sites, and numbers (1–3) indicating the regions analysed by qPCR following ChIP. The Hand2 branchial arch enhancer (green) and cardiac enhancer (yellow) are indicated. Shown in red are the ENCODE/LICR ChIP followed be sequencing (ChIP–seq) tracks for H3K4me1 and H3K27ac active enhancer markers. E1–E6, exons 1–6. See Methods for source data. c, d, H3K4me1 (c) and H3K27ac (d) modifications present at the Uph locus in E10.0 wild-type hearts are reduced in Uph KO hearts, and unchanged in UphtdTO/tdTO homozygous hearts. Values are normalized to total histone H3. e, GATA4 binding to the cardiac enhancer is reduced in E10.0 knockout hearts relative to wild type. f, ChIP using RNAPII (phosphoS2) antibody at the Hand2 transcriptional start site or gene body in wild-type, Uph KO and UphtdTO/tdTO homozygous hearts. Values normalized as a percentage of total input chromatin. Each point is one of three technical replicates of five pooled hearts for each genotype in each ChIP experiment, from one of two independent experiments; data are mean ± s.e.m.