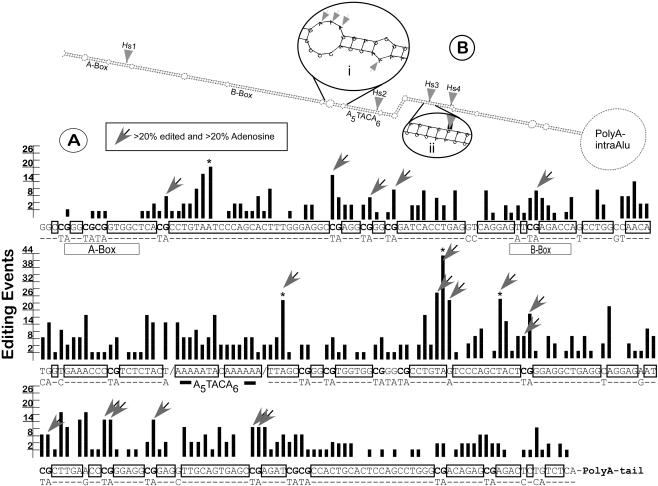
Figure 8. Sequence and Structure Preferences of Editing in Alus.
(A) The consensus sequence of 141 edited full-length Alu elements present within human Chromosome 1 transcripts with the number of editing events indicated for each sequence position (bars). Insertions and deletions present in fewer than five elements are not shown in the alignment for clarity. Bases conserved in more than 80% of the sequences are boxed. For the lesser conserved consensus positions the next most frequent base is listed below. Consensus CpG dinucleotides are in bold. Arrows indicate “high-efficiency” positions where more than 20% of adenosines present appear to be edited. Note the overlap of these positions with CpGs. Major features of Alu sequences, such as the A-Box and B-Box of Pol III and the Alu polyA sequence are labeled.
(B) A typical Alu foldback structure and its major features as discussed in the text. Arrows indicate TA hot-spot positions. The magnifications show the two typical configurations of editing sites found in Alu pairs: mismatched A/C bulges (i) and A/U base pairs (ii).