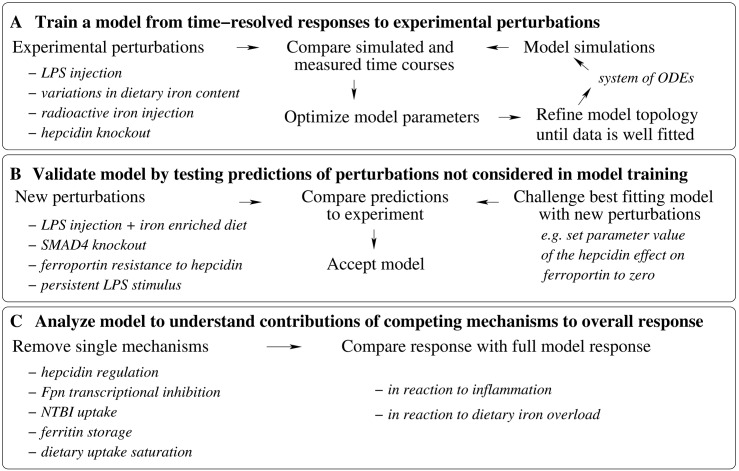
Fig 2. Summary of the different steps of the study.
A-The mathematical model based on ordinary differential equations for the time dynamics of iron pools and regulating proteins was derived from the data. Starting from a core model topology, model parameters were found by fitting simulated time courses to experimental measurements of the systems response to different perturbations. The starting model topology was successively improved by including new regulatory mechanisms (e.g. regulation of iron flows by ferroportin, regulation of ferroportin by hepcidin) and the fitting cycle repeated until the model could fit well all of the data. B-The model prediction accuracy was tested for perturbations not used in training (new experiment in this study and previously published data not used in step A). The effect of the different perturbations was simulated by changing parameter values or initial conditions of the simulations. C-The model response to inflammation and dietary iron overload was analyzed to learn the quantitative contributions of the different mechanisms included to the overall response. Several mechanisms were removed one by one and the model simulations were compared to the results of the full model.