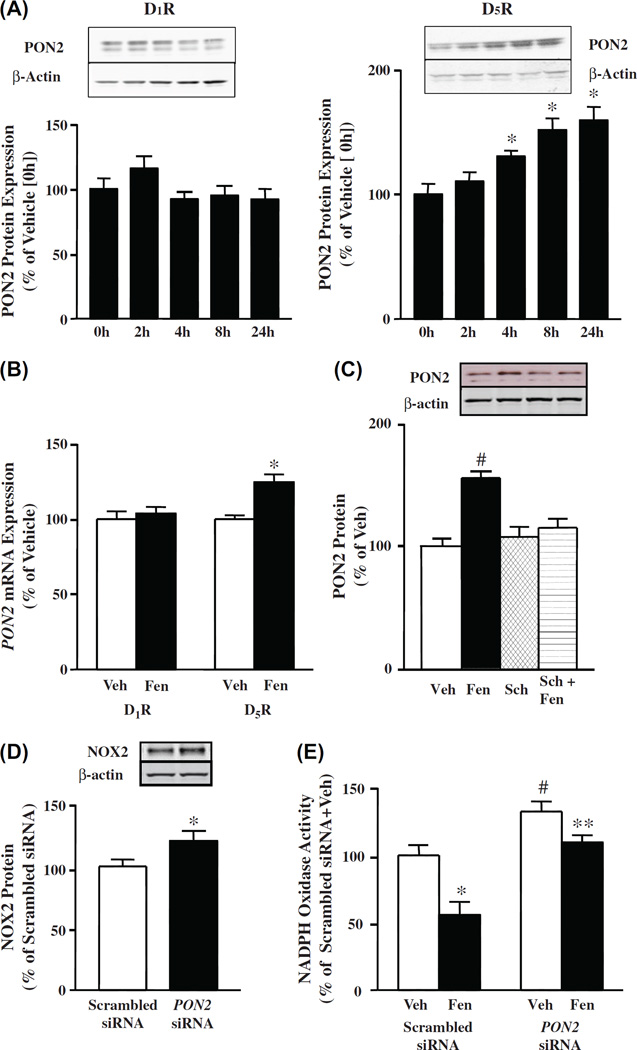
Figure 4.
Effect of long-term stimulation of D1R and D5R on PON2 protein in HEK-hD1R and HEK-hD5R cells. (A). Time course of the effect of fenoldopam on PON2 protein in HEK-hD1R and HEK-hD5R cells. The cells were treated with fenoldopam (1 µM) at varying durations (0, 2, 4, 8, 24 h) as described in “Methods”. Zero h (0 h) time was assigned a value of 100%. The data, normalized by β-actin, are shown as mean ± SEM, n = 4–6/group; *P < 0.05 vs control (0 h), one-way factorial ANOVA, Newman–Keuls test. (B). Effect of fenoldopam on PON2 mRNA expression in HEK-hD1R and HEK-hD5R cells. The cells were treated with fenoldopam (Fen, 1 µM) for 24 h. mRNA was prepared from the cell pellets, as described in “Methods”. Vehicle (Veh, control) was assigned a value of 100%. The data, normalized by GAPDH, are shown as mean ± SEM, n = 6/group, *P < 0.05 vs Veh, one-way factorial ANOVA, Newman–Keuls test. (C). Effect of fenoldopam and D1-like receptor antagonist Sch 23390 on PON2 protein in HEK-hD5R cells. The cells were treated with vehicle (Veh, which served as control, 24 h), fenoldopam (Fen, 1 µM, 24 h), Sch23390 (Sch, 1 µM, 24 h), or pretreated with Sch (1 µM, 1 h) and then incubated with Fen (1 µM, 24 h) (Sch + Fen). Veh was assigned a value of 100%. The data, normalized by β-actin, are shown as mean ± SEM, n = 4/group, #P < 0.05 vs others, one-way factorial ANOVA, Newman–Keuls test. (D). Effect of silencing PON2 on NOX2 protein in HEK-hD5R cells. Cells were transfected with PON2-siRNA (10 nM) or scrambled siRNA (10 nM) for 48 h. Scrambled siRNA was assigned a value of 100%. The data, normalized by β-actin, are shown as mean ± SEM, n = 3/group, compared with scrambled siRNA assigned a value of 100%, *P < 0.05 vs scrambled siRNA, t-test. (E). Effect of silencing of PON2 on NOX activity in HEK-hD5R cells. Cells were transfected with PON2-siRNA (10 nM) or scrambled non-silencing siRNA (10 nM) for 48 h. The cells were then treated with fenoldopam (Fen, 1 µM) or vehicle (Veh) for 24 h. Membrane NOX activity was measured in the presence of 5 µM lucigenin and 100 µM NADPH. Veh + scrambled siRNA was assigned a value of 100%. Data, normalized by protein concentration, are shown as mean ± SEM, n = 6/group, *P < 0.05 vs Veh + scrambled siRNA, #P < 0.05 vs others, **P < 0.05 vs Fen + scrambled siRNA, one-way factorial ANOVA, Newman–Keuls test.