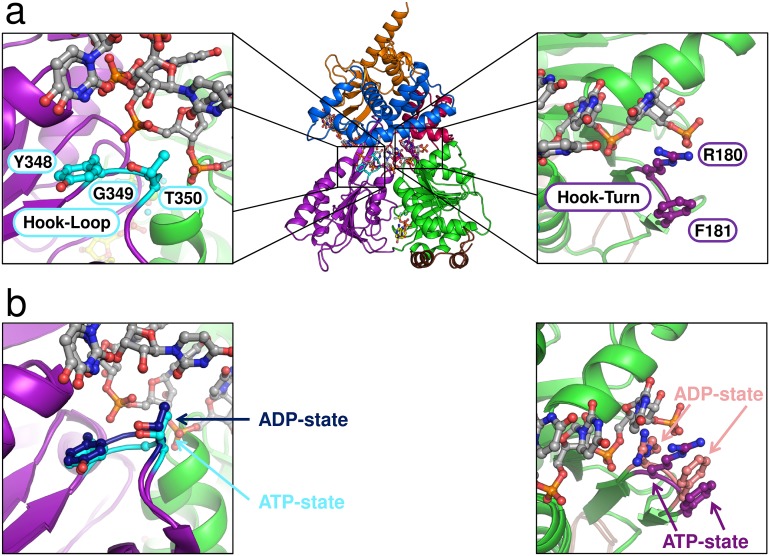
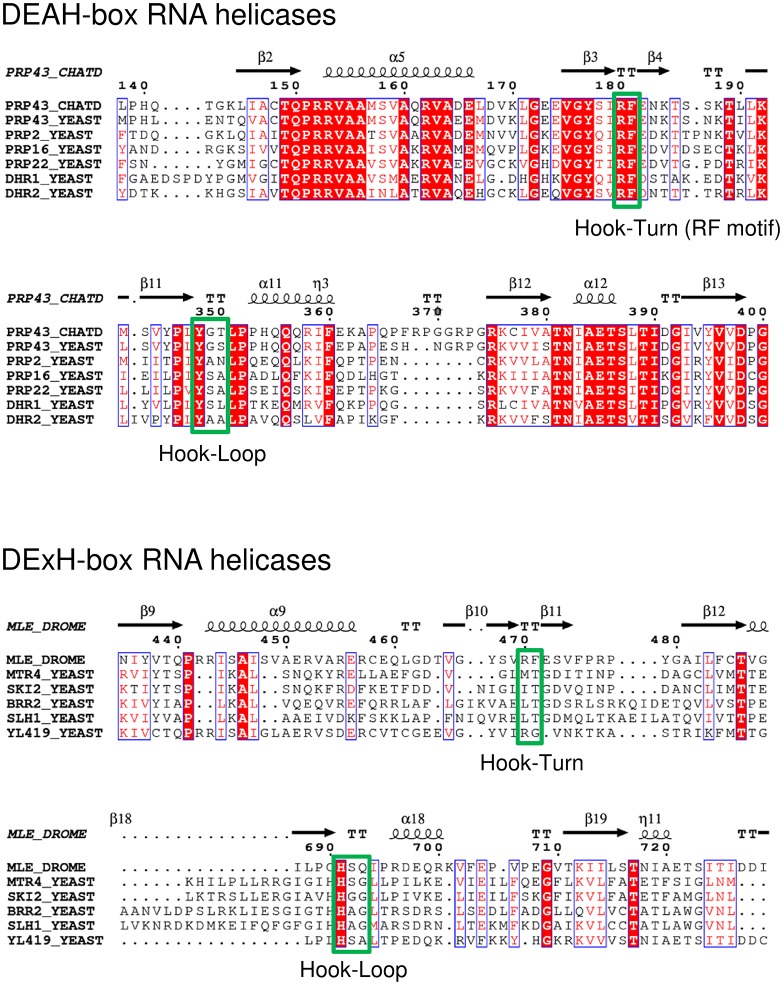
Figure 7. Position of the Hook-Turn and Hook-Loop in ctPrp43.
(a) The localization of the Hook-Turn in the RecA1 domain and of the Hook-Loop in the RecA2 domain in the ctPrp43ΔN•U7•ADP•BeF3- complex structure is shown. Domains are colored according to Figure 1a. (b) Superpositions of the RecA1 and RecA2 domains of the ctPrp43ΔN•U7•ADP•BeF3- and the ctPrp43ΔN•ADP (PDB 5d0u) complexes for the Hook-Turn and Hook-Loop, respectively. The superpositions indicate that the Hook-Loop remains in a highly similar conformation after ATP hydrolysis in contrast to the Hook-Turn which is shifted towards the RNA in the ADP-bound state.