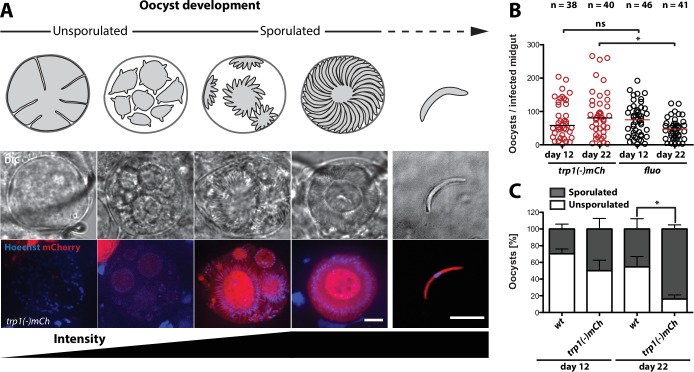
Figure 2. trp1(-) oocysts sporulate normally and persist in a sporulated state.
(A) Expression of mCherry in trp1(-)mCh parasites was only observed in sporulating oocysts and sporozoites. The developmental stage of the oocysts is depicted schematically above the images, while the increase in fluorescence intensity is schematically indicated below. Strong mCherry expression was only observed in budding or mature oocysts. Scale bar: 10 µm. (B) Oocyst numbers of infected midguts for trp1(-)mCh and wild-type parasites at day 12 and day 22 post-infection. * depicts p<0.05; one-way ANOVA followed by a Kruskal-Wallis test. Horizontal bars indicate the median. Data were generated from two (trp1(-)mCh) and three (fluo) different feeding experiments, respectively. (C) Percentages of sporulated and unsporulated oocysts in trp1(-)mCh and wild-type infected midguts at 12 and 22 days post infection. * depicts p<0.05; one-tailed Student's t-test. The mean and the SEM are shown. Data were generated from three different feeding experiments.