Fig. 2. Design, preparation, and characterization of nucleosomes with radially protruding DNA single strands.
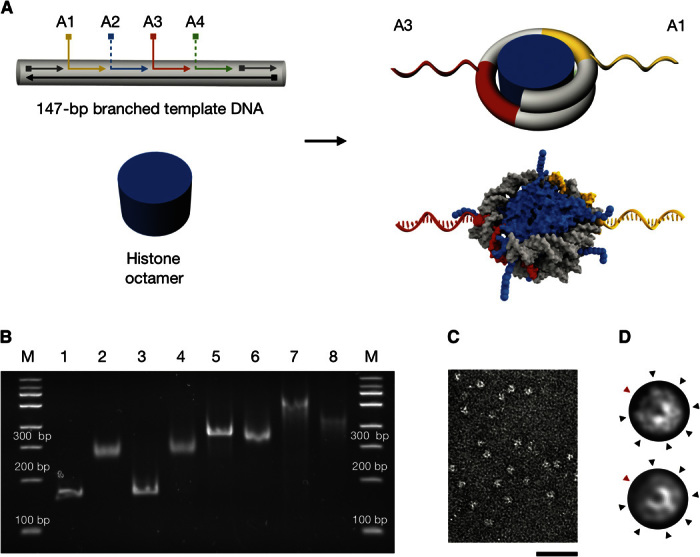
(A) Top left: Schematic of branched variants of the 601 nucleosome positioning sequence (33) with up to four (positions A1 to A4) protruding DNA single strands. Bottom left: Schematic of purified histone octamer. Right: Schematics of nucleosomes with radially protruding DNA single strands at positions A1 and A3 that are produced by salt gradient dialysis from the components on the left. The bottom schematic is based on 3MVD.pdb (4). (B) Native ethidium bromide–stained 4.5% polyacrylamide gel electrophoresis of various samples: lane 1, continuous template DNA; lane 2, nucleosomes assembled using continuous template DNA as in lane 1; lane 3, template DNA with four nicks at positions A1 to A4; lane 4, nucleosomes assembled using nicked template DNA as in lane 3; lane 5, template DNA with two protruding single strands at positions A1 and A3 [see (A)]; lane 6, nucleosomes assembled using template DNA with two protruding single strands as in lane 5; lane 7, template DNA with four protruding single strands at positions A1 to A4; lane 8, nucleosomes assembled using the template DNA as in lane 7. Nucleosomes were assembled by salt gradient dialysis reconstitution with Drosophila embryo histones. See note S1 for detailed protocol. (C) Representative electron micrograph of nucleosomes with two protruding DNA single strands (A1 and A3) and recombinant tailless histones from X. laevis. Scale bar, 50 nm. (D) Projection of a nucleosome crystal structure (3MVD.pdb) (top) and average electron micrograph from nucleosomes shown in (C) (bottom). See fig. S4 for additional data. Black arrowheads indicate radial intensity signatures stemming from grooves in the DNA template. Red arrowheads indicate the dyad axis in the arc segment having only one DNA turn.
