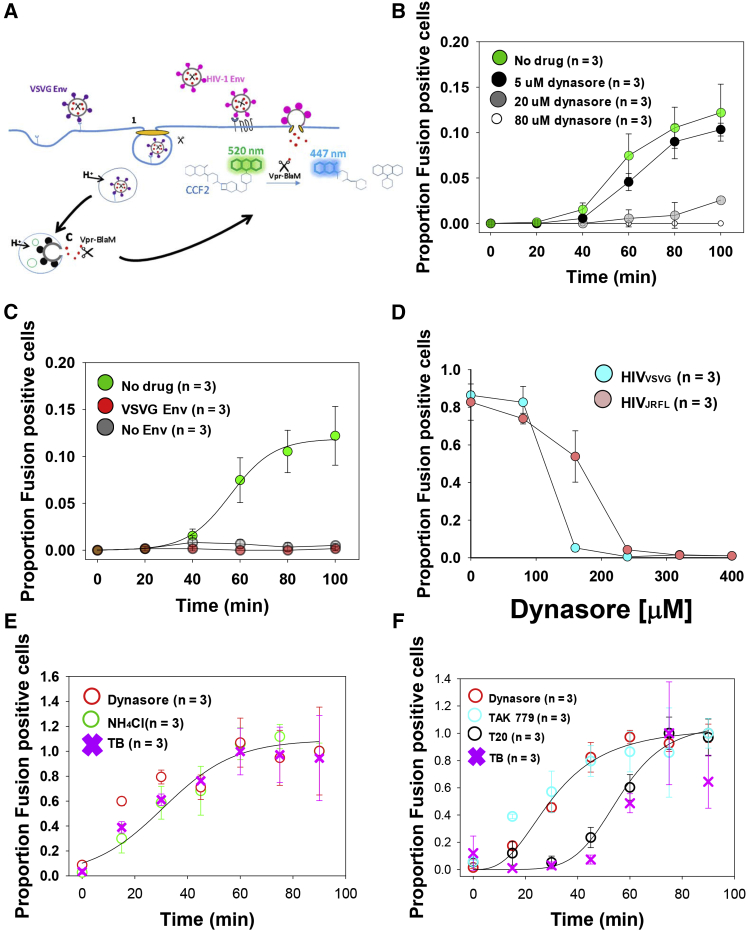
Figure 1.
HIV-1 Fusion Kinetics Is Dynamin-2 Dependent in Both CD4 T Cells and TZM-bl Cells
(A) Cartoon depicting the BlaM assay. Upon virion fusion and capsid release, the Vpr-BlaM chimera recognizes a FRET reporter (CCF2) that changes color (green to blue) upon cleavage.
(B) Real-time BlaM was applied using HIV-1 virions packaging the Vpr-β-Lactamase chimera and pseudotyped with HXB2 Env on primary CD4 T cells at different concentrations of dynasore: 0 μM (green dots), 5 μM (black dots), 20 μM (gray dots), and 80 μM (white dots). The proportion of fusion positive cells versus total number of cells is shown (y axis) versus time, in min (x axis).
(C) HIV-1/Vpr-β-Lactamase virions pseudotyped with VSVG turned out not to be fusogenic (red dots) showing the same behavior as HIV1/Vpr-β-Lactamase bald particles (without Env, black dots).
(D) HIV-1 virions packaging the Vpr-β-Lactamase chimera and pseudotyped with either VSV-G (cyan dots) or JR-FL (orange dots) were exposed to TZM-bl cells with different concentrations of dynasore (0, 100, 180, 260, 340, and 400 μM) and endpoint BlaM (as defined in Experimental Procedures) was applied. Higher concentrations of dynasore were required to fully inhibit HIVJRFL (240 μM) as compared with HIVVSVG (180 μM).
(E) Time-of-addition BlaM kinetics without spinoculation protocols on HIVVSV-G virions using three different blocks: 400 μM dynasore (open red dots), temperature block (pink crosses), and NH4Cl (open green dots). All of the kinetics turned out to be very similar. The normalized proportion of fusion positive cells versus total number of cells is shown (y axis) versus time, in min (x axis).
(F) Time-of-addition BlaM kinetics without spinoculation protocols on HIVJRFL virions using four different blocks: TAK 779 (open blue dots), dynasore (open red dots), T20 (open black dots), and temperature block (pink crosses). The normalized proportion of fusion positive cells versus total number of cells is shown (y axis) versus time, in min (x axis). In all cases, the error bars represent the SD calculated from three independent experiments.