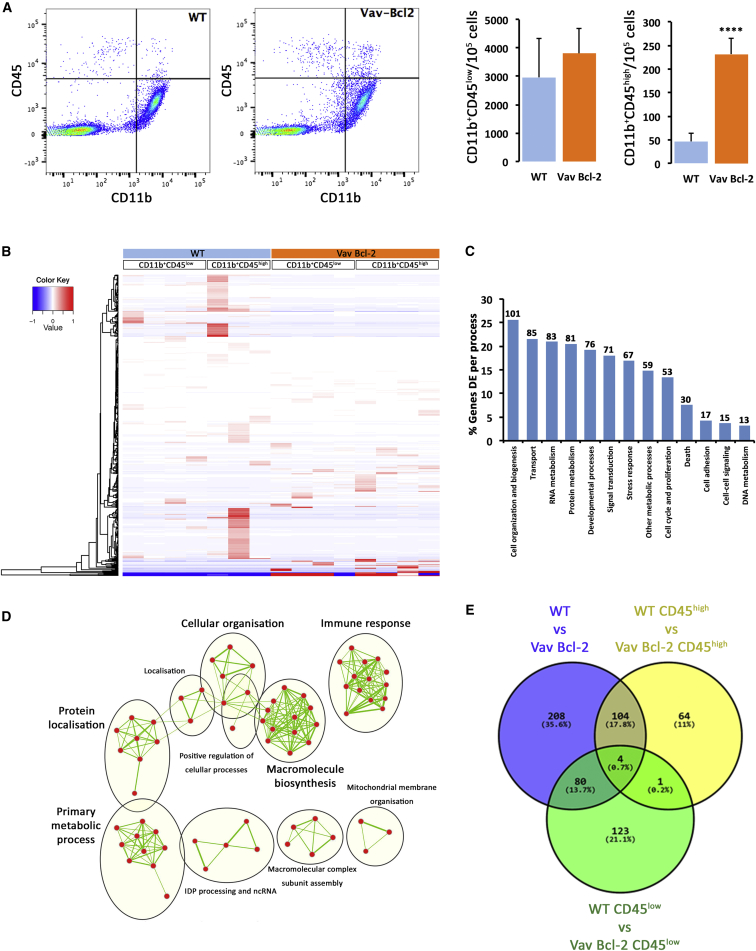
Figure 6.
Transcriptomic Profiling of Microglia from WT and Vav-Bcl2 Mice
(A) Flow cytometry analysis and sorting of microglia from WT and Vav-Bcl2 mice. Crosshair in flow cytometry analysis and sorting plots shows gating parameters used to define CD11b+CD45low and CD11b+CD45high subpopulations and subsequent sorting. Statistical differences: ∗∗∗∗p < 0.0001. Data were analyzed with a t test (A).
(B) Heatmap representation of genes showing a significant (p < 0.01; >10-fold change) change in Vav-Bcl2 versus WT microglia (combined CD45+). Clustering of genes by expression profile is shown on the left.
(C) Clustered representation (GO Slim) of GO processes significantly altered in Vav-Bcl2 compared to WT microglia. Number of genes altered per cluster is shown on top of the bars.
(D) Enrichment map of GO terms, where red nodes represent GO terms and green edges represent shared genes (thicker lines indicate more shared genes).
(E) Venn diagram representing the intersection of the transcriptional variability observed when comparing total (blue), CD45low (green), or CD45high (yellow) Vav-Bcl2 to WT microglia.