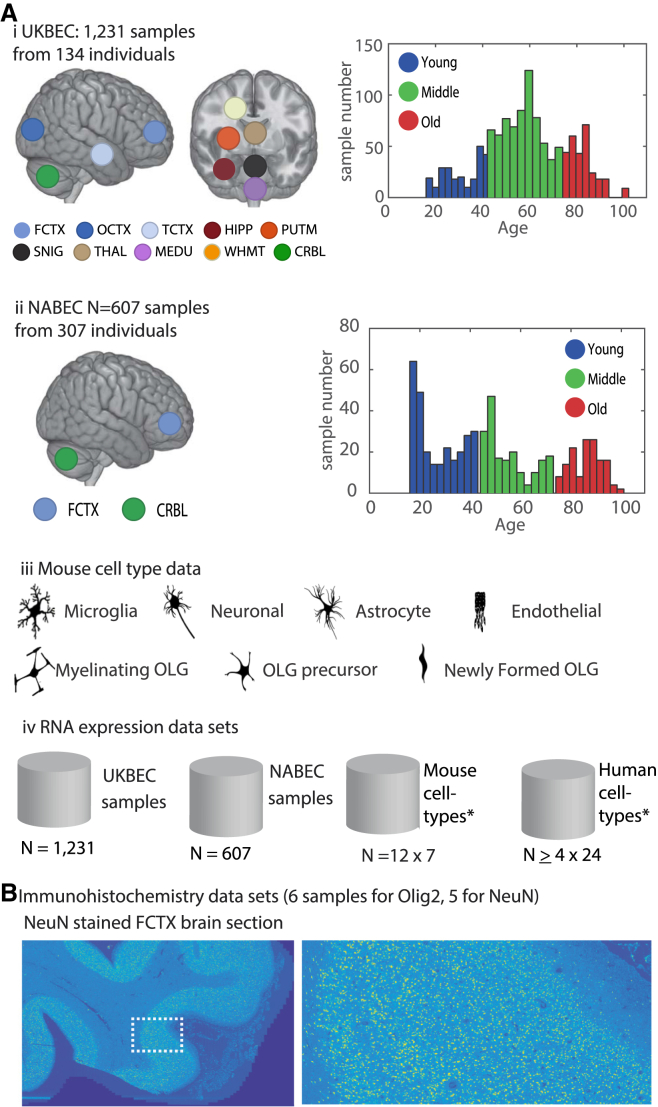
Figure 1.
Analyzed Samples and Datasets
(A) The samples of the UKBEC and NABEC datasets were divided into three age groups each (young: 16–44, middle: 45–74, old: ≥75). (i) The main analyzed dataset (UKBEC) is composed of 1,231 brain samples interrogated by exon microarrays, from brain samples of 134 individuals from 16 to 102 years old and up to ten brain regions each. The brain regions included both cortical and sub-cortical regions, specifically: the frontal cortex (FCTX), temporal cortex (TCTX), occipital cortex (OCTX), intralobular white matter (WHMT), cerebellum (CRBL), substantia nigra (SNIG), putamen (PUTM), thalamus (THAL), hippocampus (HIPP), and medulla (MEDU) for UKBEC and the FCTX and CRBL for NABEC. (ii) The independent (NABEC) dataset of brain samples from FCTX and CRBL 307 individuals (16–101 years old). (iii) In addition, seven cell types were identified based on analysis of available RNA-seq data from mice cortex (http://web.stanford.edu/group/barres_lab/brain_rnaseq.html). (iv) A summary of all expression data used in this study. The total number of samples described in (i)–(iii) is listed, as well as the human RNA-seq analysis of 24 CNS human cell types (Table S7) (http://web.stanford.edu/group/barres_lab/brainseqMariko/brainseq2.html).
(B) High-resolution immunohistochemical imaging dataset was produced from samples of young and three old FCTX from the UKBEC cohort, following staining by OLIG2 antibody and computational analysis for the quantification of the OLG cell population. Staining by NeuN of FCTX sections from the same brain samples followed by targeted computational analysis was conducted for quantification of the neuronal cell population (an example of one of the NeuN stained sections is shown on the right, in the zoomed-in view of the area marked on the left-hand side). OLG, oligodendrocyte.