Figure 3.
MLL-AF4 Spreading Marks a Subset of Highly Expressed Genes
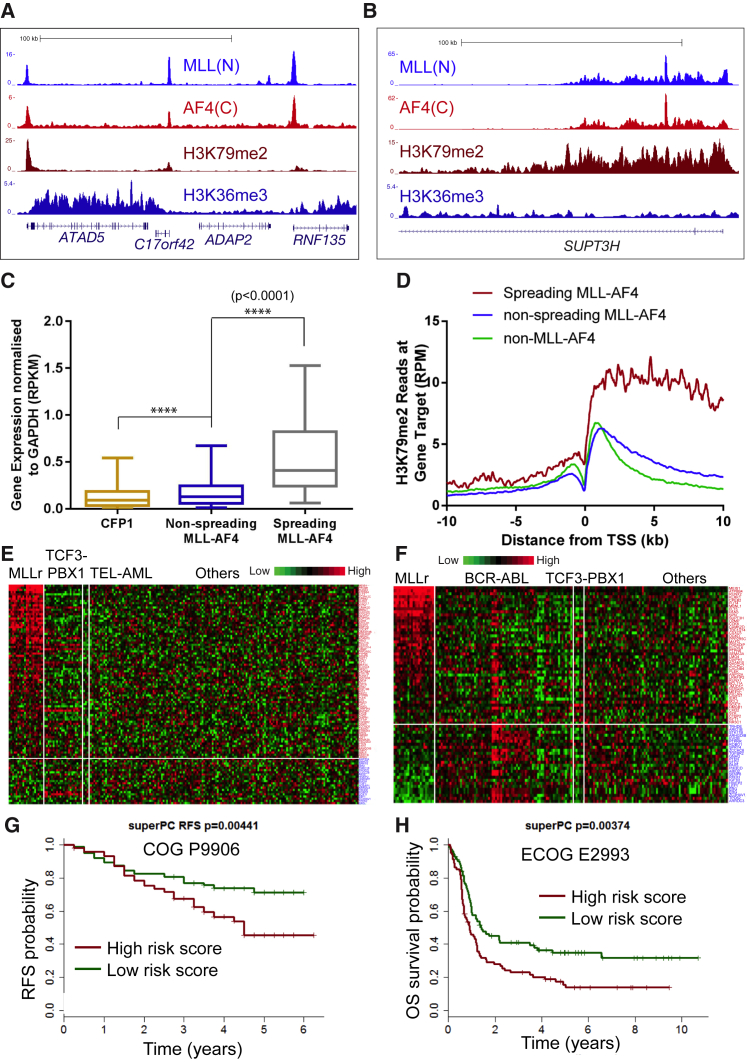
(A and B) Example ChIP-seq tracks showing promoter-restricted (A) or spreading (B) of MLL-AF4, H3K79me2, and H3K36me3 in SEM cells.
(C) Box-and-whisker plot showing the median and interquartile (IQ) range of gene expression of spreading MLL-AF4 gene targets (n = 149) compared to non-spreading MLL-AF4 targets (n = 2,878) and CFP1 targets (n = 6,147). Gene expression, normalized to GAPDH expression, is derived from four biological replicates of nascent RNA-seq in SEM cells. ∗∗∗∗p < 0.0001, two-tailed Mann-Whitney U test.
(D) Composite binding plot of H3K79me2 ChIP-seq reads at the TSS of gene targets of spreading MLL-AF4 (red), non-spreading MLL-AF4 (blue), and non-MLL-AF4 targets that are marked by H3K79me2 (green).
(E and F) Heatmap expression data showing overexpression of 79% (E, COG P9906 patients [Harvey et al., 2010]) or 64% (F, ECOG 2993 patients [Geng et al., 2012]) of SEM spreading targets in MLL patients (MLLr) compared to the ALL patient subsets indicated.
(G and H) Super-PC analysis (Bair and Tibshirani, 2004) using the spreading-gene target list showing relapse-free survival (RFS) of ALL patients (G, COG P9906 [Harvey et al., 2010]) and overall survival (OS) of ALL patients (H, ECOG 2993 [Geng et al., 2012]) classified by either high- or low-risk scores computed using the spreading MLL-AF4 gene targets in a super-PC model; see Supplemental Experimental Procedures, Survival Analysis, for details.
See also Figure S3.