Figure 4.
MLL-FP Spreading Occurs in Multiple in MLL Leukemias
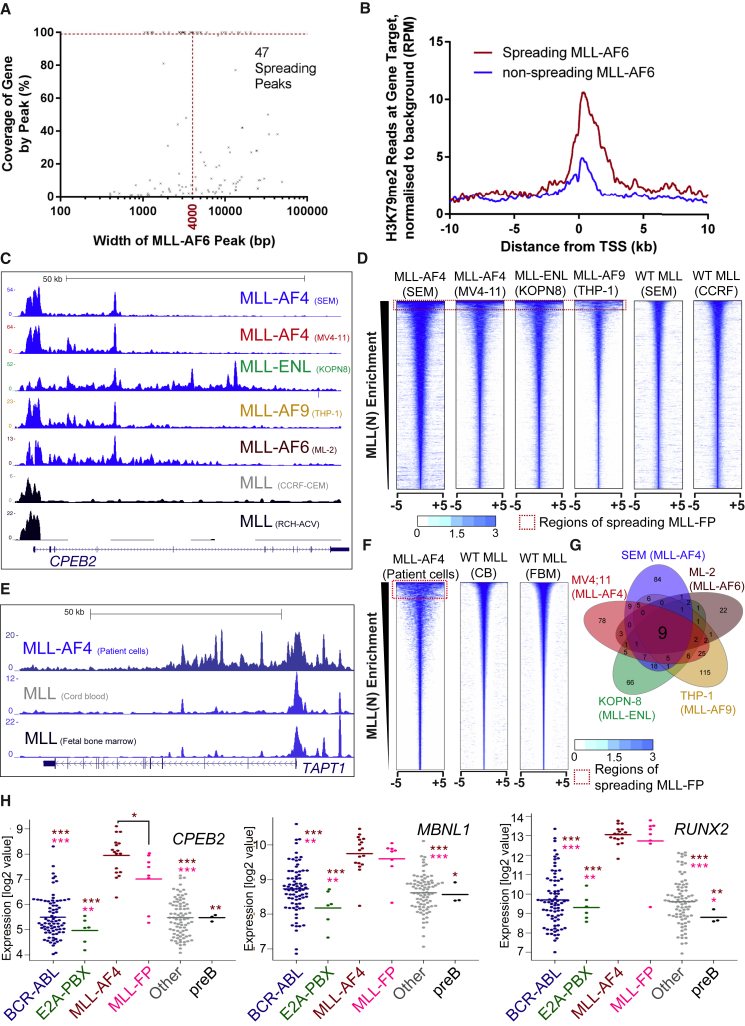
(A) Spreading MLL-AF6 peaks were defined as peaks that extend greater than 4 kb from the TSS into the gene body without going beyond the end of the gene. Using these criteria, 47 spreading MLL-AF6 peaks were identified in ML-2 cells (Table S2).
(B) Composite binding plot of H3K79me2 ChIP-seq reads at the TSS of gene targets of spreading MLL-AF6 (red) and non-spreading MLL-AF6 (blue) in ML-2 cells.
(C) Example ChIP-seq tracks of MLL(N) in MLL-FP and germline MLL cell lines.
(D) Heatmaps of MLL(N) ChIP-seq reads from different MLL-FP cell lines as well as wild-type MLL in SEM cells and in non-MLLr cell lines. Red dotted line indicates spreading across a 10-kb window. Scale bar represents tags per base pair per 107 reads.
(E) Example ChIP-seq tracks of MLL(N) showing spreading in MLL-AF4 patient cells (top) compared to wild-type MLL in mononuclear cells derived from cord blood (middle) and fetal bone marrow (bottom).
(F) Heatmaps showing MLL(N) ChIP-seq reads from the experiments in (E); scale and red line as in (D).
(G) Venn diagram showing the overlap between gene targets of spreading MLL(N) ChIP-seq several MLLr cell lines.
(H) CPEB2, MBNL1, and RUNX2 are overexpressed in MLL-AF4 and other MLL-FP patients compared to different patient samples and normal pre-B cells. Each dot indicates an individual patient sample. Data are taken from an ECOG E2993 clinical trial (Geng et al., 2012). Dark red asterisk (∗) indicates a significant difference compared to MLL-AF4, and pink asterisk (∗) indicates a significant difference compared to the MLL-FP group (which includes MLL-ENL [6], MLL-AF9 [1], and MLL-EPS15 [1]). ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05. A two-tailed Wilcoxon test was used to calculate p values, and p values for the different comparisons are listed in Table S5.
See also Figures S4 and S5.