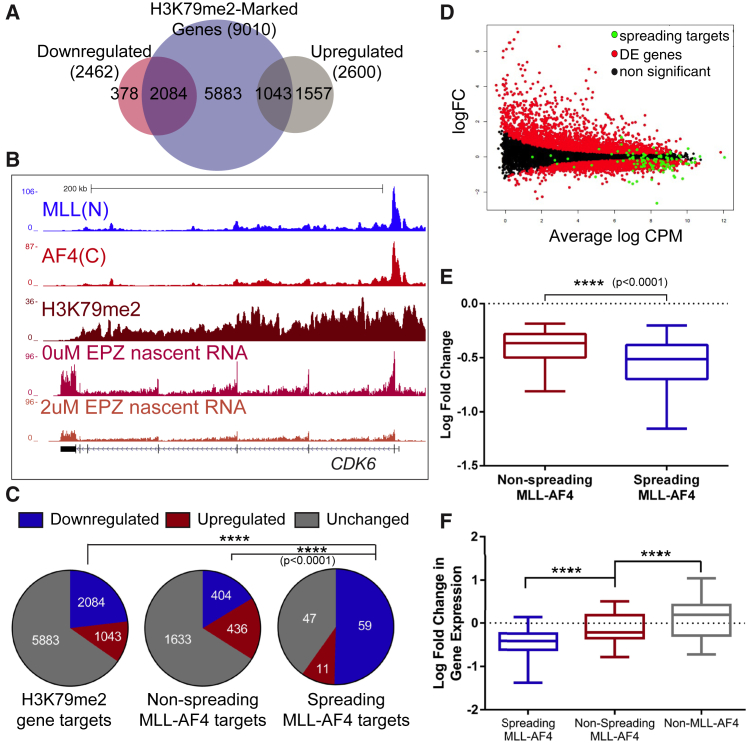
Figure 6.
Spreading MLL-AF4 Targets Show Increased Sensitivity to DOT1L Inhibition
(A) Venn diagram showing an overlap between H3K79me2-marked genes and upregulated and downregulated genes in SEM cells following treatment with 2 μM EPZ-5676.
(B) Example ChIP-seq tracks at CDK6 and nascent RNA-seq in control (0 μM) and 2 μM EPZ-5676-treated SEM cells.
(C) Pie charts showing the proportion of genes that are significantly downregulated (blue), upregulated (red), or remain unchanged (gray), among H3K79me2-marked genes (left), non-spreading MLL-AF4 gene targets (center), and spreading MLL-AF4 gene targets (right), following treatment of SEM cells with 2 μM EPZ-5676. ∗∗∗∗p < 0.0001, Fisher’s exact test.
(D) Smear plot showing the fold change in gene expression of all genes in SEM cells following treatment with 2 μM EPZ-5676 compared to their expression level (CPM). Black, non-significant change in gene expression; red, differentially expressed gene; green, spreading MLL-AF4 gene targets.
(E) Box-and-whisker plot showing the median and IQ range of fold change in expression of all significantly downregulated gene targets of non-spreading MLL-AF4 (red) compared to spreading MLL-AF4 (blue), after 2 μM EPZ-5676 treatment in SEM cells. ∗∗∗∗p < 0.0001, Mann-Whitney U test.
(F) Box-and-whisker plot showing the median and IQ range of fold change in expression of all significantly affected spreading MLL-AF4 (blue), non-spreading MLL-AF4 (red), and non-MLL-AF4 gene targets following siRNA-mediated knockdown of MLL-AF4 in SEM cells. ∗∗∗∗p < 0.0001, Mann-Whitney U test.
See also Figure S7.