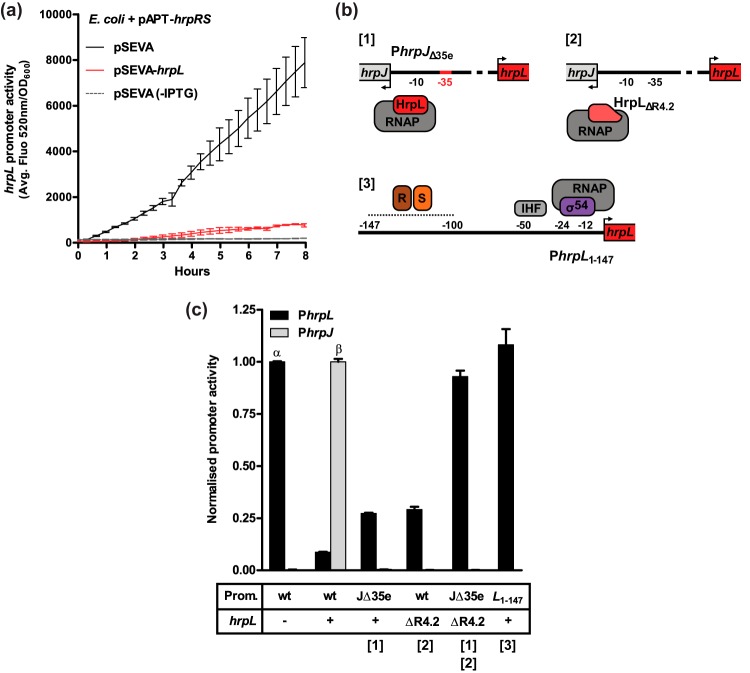
FIG 3 .
(a) hrpL promoter activity in an E. coli s17λpir strain carrying the pBBR1-PhrpL-gfp reporter in LB medium. Heterologous expression of hrpL from pSEVA614-hrpL (red) and hrpRS from pAPT-hrpRS was induced with 0.1 mM IPTG. The empty pSEVA614 vector (black) was used to control for plasmid load. HrpRS-independent PhrpL activity was measured in the absence of IPTG (dashed gray line). Error bars represent SEM of results of 3 biological replicates. (b) Mutations disrupting the association of the RNAP-HrpL with PhrpJ DNA. [1] Substitution of the hrp-box −35 element (GGAACT > AACACT). [2] Truncation of HrpL DNA-binding region 4.2. [3] Exclusion of PhrpJ using a minimal promoter sequence sufficient for PhrpL activity (−147 to +1). (c) The relative activity levels of PhrpL and PhrpJ seen when HrpL DNA-binding function was disrupted. The expression states of hrpL and the cis- and trans-acting mutations acting under each condition are tabulated below the graph. PhrpL (GFP) and PhrpJ (RFP) activities were measured simultaneously under all conditions using the reporters pBBR1-rfp-PhrpL-gfp (wt) or pBBR1-rfp-PhrpL(Δ35e)-gfp (JΔ35e), with the exception of the minimal pBBR1-PhrpL(147)-gfp reporter (L1-147; far-right column). hrpL (+) or hrpLΔR4.2 (ΔR4.2) were expressed from the pSEVA614 plasmid, the empty vector being used as a proxy for the ΔhrpL condition (−). HrpRS proteins were expressed from pAPT-hrpRS under all conditions. Promoter (Prom.) activities given are normalized relative to the maximum activity state for each condition after 8 h of growth in LB as follows: for PhrpL, empty pSEVA614/wild-type promoter (α); for PhrpJ, pSEVA614-hrpL/wild-type promoter (β). Error bars represent SEM of results of 3 biological replicates.