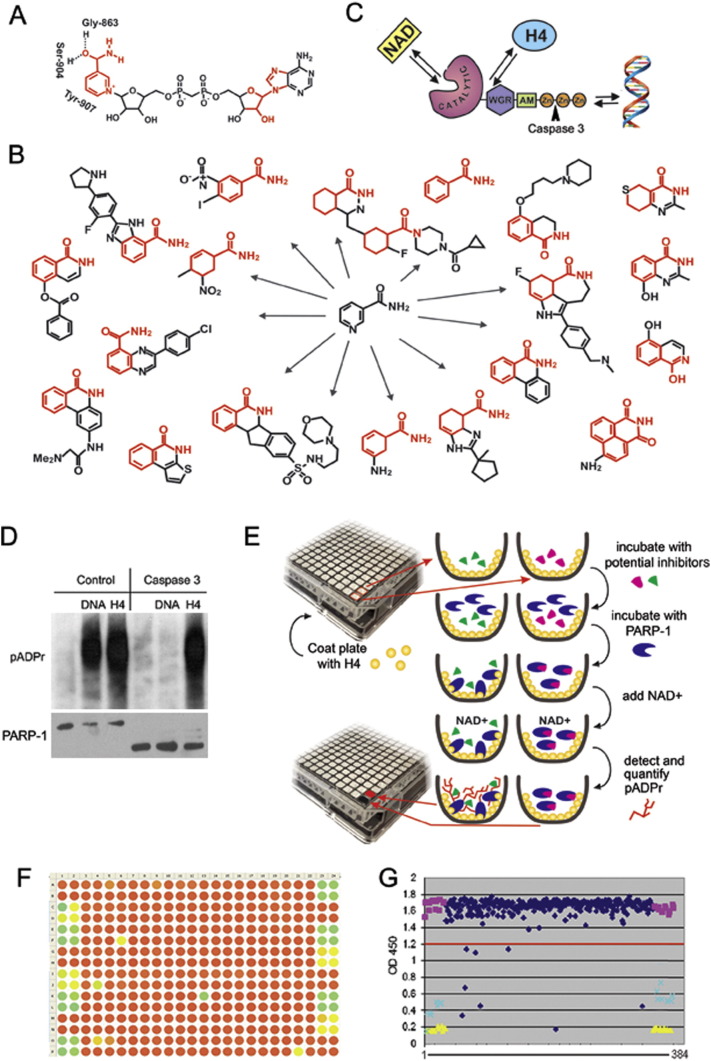
Fig. 1.
Designing a new screening strategy to identify PARP-1 inhibitors. A. PARP-1 binds NAD + by NAD-binding pocket organized by three amino acids, including Gly-863, Ser-904 and Tyr-907, which mostly interact with the nicotinamide part of NAD. The parts of NAD used to develop PARP-1 inhibitors are shown in red. B. Most current PARP-1 inhibitors are developed from nicotinamide pharmacophore. C. Three ways of PARP-1 regulation: 1) competition with NAD for binding, 2) disruption of PARP-1 interaction with histones and 3) obstruction of binding with DNA. Arrowhead shows site of PARP-1 digestion by Caspase 3, which cleaves off DNA binding Zn-fingers of PARP-1, thus abolishing DNA-dependent PARP-1 activation. D. Interaction with the purified core histone H4 activates PARP1 in a DNA-independent manner. Full-length PARP-1 protein (left) and PARP1 protein cleaved by Caspase 3 (right) were preincubated with randomly broken DNA or core histone H4, followed by mixing with NAD. The product of PARP-1 enzymatic activity, poly(ADP-ribose), was detected after PAGE on a Western blot using anti-pADPr antibody. These data clearly demonstrate that the DNA-binding domain of PARP1 (Zn-fingers I and II) is not required for histone-dependent PARP1 activation. E. Schematic representation of the pipeline used to identify PARP-1 inhibitors. F-G. Data were visualized in a colour-coded table representing the 384-well plate in which potential inhibitors could be identified as green or yellow circles corresponding to wells that had minimal pADPr signal (F) or on a graph representing relative numerical value of this signal when compared to positive (yellow) or negative (purple) controls (G).