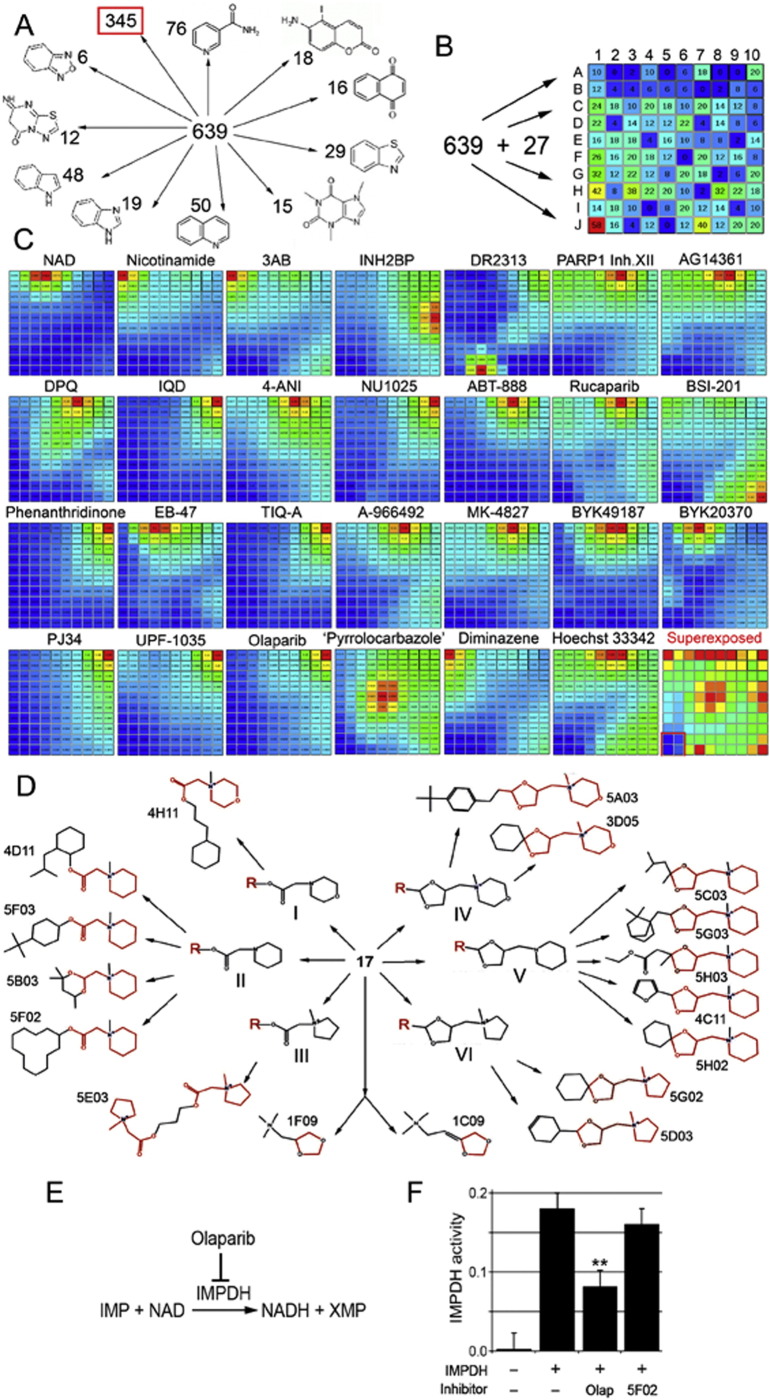
Fig. 2.
Identifying non-NAD-like small molecules inhibiting PARP-1 protein. A. Sorting out new PARP-1 inhibitors based on the presence of an obvious structural core, similar to known biologically active molecules. Eleven subgroups were identified. Structural cores and numbers of molecules falling in each group are indicated. B. Sorting out 639 new plus 27 known PARP-1 inhibitors and NAD based on 3D fingerprints, using the Canvas, ver. 1.6, program. Based on similarity of fingerprints, compounds were sorted to a 2D matrix containing 100 cells. The number of small molecules with all their 3D isomers in each cell is indicated. C. Comparison of molecules sorted in the matrix with each known PARP-1 inhibitor and NAD. Similarity is illustrated by heat map. Red corresponds to highest similarity and blue to absence of similarity. Name of inhibitor is indicated above the map. Bottom-right square represents a superimposure of 27 heat maps and reveals the area of matrix (labeled with red border) containing non-NAD-like compounds. D. Molecular structures of new PARP-1 inhibitors. Structural cores: I - 2-(N-methylmorpholino) acetate; II - 2-(N-methylpiperidin-1-yl)acetate; III - 2-(N-methylpyrrolidine-1-yl)acetate; IV - 1-((1,3-dioxolane-4-yl)methyl)N-methylmorpholino; V - 1-((1,3-dioxolane-4-yl)methyl) piperidine; VI - 1-((1,3-dioxolane-4-yl)methyl) N-methylpyrrolidine. E. Schematic illustration of IMPDH2 catalyzing reaction. F. Non-NAD-like inhibitors do not disrupt IMPDH2 activity. Graph showing IMPDH specific- activity in the IMPDH reaction with/without PARP-1 inhibitors. Column1: no recombinant human IMPDH2 added in the reaction. Column2: human IMPDH2 with 2 ul DMSO in the reaction. Column3: human IMPDH2 with 2 mM Olaparib in the reaction. Column 4: human IMPDH2 with 2 mM 5F02 in the reaction. **: P < 0.01.