Abstract
Infection with one of the four dengue virus serotypes (DENV1-4) presumably leads to lifelong immunity against the infecting serotype but not against heterotypic reinfection, resulting in a greater risk of developing Dengue Hemorrhagic Fever/Dengue Shock Syndrome (DHF/DSS) during secondary infection. Both antibodies and T cell responses have been implicated in DHF/DSS pathogenesis. According to the T cell-based hypothesis termed “original antigenic sin,” secondary DENV infection is dominated by non-protective, cross-reactive T cells that elicit an aberrant immune response. The goal of our study was to compare the roles of serotype-specific and cross-reactive T cells in protection vs. pathogenesis during DENV infection in vivo. Specifically, we utilized IFN-α/βR−/− HLA*B0702 transgenic mice in the context of peptide vaccination with relevant human CD8 T cell epitopes. IFN-α/βR−/− HLA*B0702 transgenic mice were immunized with DENV serotype 2 (DENV2)-specific epitopes or variants found in any of the other three serotypes (DENV1, DENV3 or DENV4), followed by challenge with DENV. Although cross-reactive T cell responses were lower than responses elicited by serotype-specific T cells, immunization with either serotype-specific or variant peptide epitopes enhanced viral clearance, demonstrating that both serotype-specific and cross-reactive T cells can contribute to protection in vivo against DENV infection.
Abbreviations: Ab, antibody; ADE, antibody dependent enhancement; DENV, dengue virus; DHF, dengue hemorrhagic fever; DSS, dengue shock syndrome; HLA, human leukocyte antigen; ICS, intracellular cytokine staining; IFN, interferon; NS, non-structural; PBMC, Peripheral Blood Mononuclear Cells
Keywords: Dengue, Cross-reactivity, T cells, Vaccination
Highlights
-
•
Serotype-cross-reactive CD8 T cells elicit a polyfunctional immune response similar to the response elicited by serotype-specific CD8 T cells.
-
•
Serotype cross-reactive CD8 T cells play a role in protection against DENV infection in a HLA-B*0702 Transgenic IFN-α/βR−/− mouse model.
There are four major subtypes (serotypes) of the mosquito-borne Dengue virus. Infection with a first serotype is generally asymptomatic, but secondary infection with a different serotype is capable of causing severe disease. T cells previously exposed to a first serotype and which produce an immune response to a second serotype are said to be cross-reactive. Using a mouse model engineered with human T cell features, we characterized the cross-reactive T cell response to live dengue virus serotypes and viral protein fragments. Our results suggested cross-reactive T cells contribute to control of and protection against infection by a second dengue serotype, rather than leading to more severe disease.
1. Introduction
Dengue virus (DENV), a member of the Flaviviridae family, is the most prevalent arthropod-borne virus in the world. The incidence of DENV infections in endemic areas has increased 30-fold in the past 50 years due to demographic changes, urbanization and globalization (Halstead, 2007, Guzman et al., 2010). New estimates report 390 million infections per year, with 96 million being symptomatic, of which > 500,000 are reported as severe forms of dengue (Bhatt et al., 2013).
DENV is a positive sense, single-stranded RNA virus and its genome is translated as a single poly-protein that is cleaved into three structural (capsid (C), pre-membrane (PrM), and envelope (E)) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (Halstead, 2007). Infection with one of the four DENV serotypes can cause a spectrum of illnesses that range from dengue fever (DF) to severe forms of dengue, previously known as dengue hemorrhagic fever/dengue shock syndrome (DHF/DSS) (Malavige and Ogg, 2012, Jayaratne et al., 2012).
Severe dengue disease is characterized by thrombocytopenia, elevated hematocrit and cytokine levels, increased vascular permeability, and hemorrhagic manifestations; it can ultimately lead to death (Halstead, 2012). The mechanisms involved in the pathogenesis of the severe forms of dengue infection remain poorly understood. Infection with one serotype confers life-long immunity against homotypic reinfection; however, individuals re-infected with a different serotype are prone to developing severe disease (Halstead, 2007). Two main hypotheses implicating the host immune response have been proposed to explain dengue pathogenesis in individuals with heterotypic secondary infection. According to the antibody dependent enhancement of infection (ADE) hypothesis, non-neutralizing antibodies from a previous infection enhance viral entry via Fcγ receptor (FcγR)-bearing cells upon reinfection. Studies using mouse models of experimental DENV infection formally demonstrated ADE in vivo, providing support for the ADE hypothesis (Zellweger et al., 2010, Balsitis et al., 2010). In contrast to ADE, the “original T cell antigenic sin” hypothesis focuses on the T cell response (Rothman et al., 2014). It postulates that memory cross-reactive T cells are preferentially activated during secondary infection, resulting in ineffective control of the infecting serotype and impairment of viral clearance (Mongkolsapaya et al., 2003, Bashyam et al., 2006). To date, direct evidence in support of the original T cell antigenic sin hypothesis is lacking. On the contrary, increasing number of studies using mouse models have shown a direct contribution of T cells in protection against DENV infection (Yauch et al., 2009, Yauch et al., 2010, Prestwood et al., 2012, Zellweger et al., 2013, Zellweger et al., 2014, Zellweger et al., 2015). In particular, we recently demonstrated that CD8 T cells could directly contribute to protection against heterotypic reinfection in mice (Zellweger et al., 2015). Consistent with these mouse findings, recent studies using DENV-exposed blood donors from a hyperendemic country support an HLA-linked protective role for T cells against DENV infection in humans (Weiskopf et al., 2013, Weiskopf et al., 2015a). Moreover, studies analyzing the T cell response in general populations from two different hyperendemic countries, Sri Lanka and Nicaragua, showed that although the T cell specificity was skewed towards conserved regions, this was not associated with impairment of the actual immune response (Weiskopf et al., 2013, Weiskopf et al., 2015b). However, studies examining T cell cross-reactivity in humans can be challenging to conduct, as the viral strain, sequence, or interval between infections is unknown.
To perform mechanistic studies on the T cell response to DENV, a manipulable and tractable animal model is required in which the exact infecting viral strain, as well as the order and times of infection, are known. Wild-type mice are highly resistant to infection with DENV, as the virus is able to block type I Interferon (IFN) receptor signaling in human but not murine cells (Ashour et al., 2010, Yu et al., 2012, Aguirre et al., 2012). The antiviral IFN response must be disrupted in mice to make them susceptible to DENV infection. Therefore, we previously developed a model of DENV infection in IFN-α/βR−/− mice lacking the type I IFN receptor (Perry et al., 2009, Zellweger et al., 2010). Our group has used this mouse model to identify protective roles for T cells in the context of various DENV infection settings, including heterotypic reinfection (Yauch et al., 2009, Yauch et al., 2010, Zellweger et al., 2010, Zellweger et al., 2015, Zellweger et al., 2014, Zellweger et al., 2013).
To link the findings obtained from the IFN-α/βR−/− mouse model studies to the human situation, we have also developed an IFN-α/βR−/− HLA transgenic mouse model. This mouse model has been used to map the DENV2- and DENV3-specific T cell response restricted by HLA B*0702 (Weiskopf et al., 2011, Weiskopf et al., 2014), which is associated with high response frequency and magnitude and decreased susceptibility to severe dengue disease in humans (Weiskopf et al., 2013). Additionally, the HLA B*0702-transgenic IFN-α/βR−/− mouse model has been validated by following observations: (i) The epitopes identified were also recognized by human Peripheral Blood Mononuclear Cells (PBMC) from DENV-exposed donors (Weiskopf et al., 2011, Weiskopf et al., 2014); (ii) A dominance of HLA B*0702-restricted response was detected in both mice and human peripheral blood mononuclear cell (PBMC) donors (Weiskopf et al., 2013); and (iii), The finding that CD8 T cell response targets both structural and NS proteins in DENV3 but only NS proteins in DENV2 was observed in both mice and humans (Weiskopf et al., 2015b, Weiskopf et al., 2015c).
In the present study, we explore the role of cross-reactive T cells in vivo during DENV infection using the HLA-B*0702-transgenic IFN-α/βR−/− mouse model. We show that HLA-B*0702-transgenic IFN-α/βR−/− mice infected with DENV2 mount a poly-functional antiviral immune response. In particular, CD8 T cells from DENV2-infected mice produce IFNγ, TNFα, or both cytokines when re-stimulated with DENV2 or variant peptides from other serotypes, DENV1/3/4. Although cross-reactive T cells elicit a weaker T cell response in vitro, immunization with cross-reactive peptides confers protection against DENV infection by reducing viral load. Our results do not provide support for “original T cell antigenic sin;” instead, they demonstrate that cross-reactive T cells can play a protective role against DENV infection.
2. Methods
2.1. Mice and Viral Strains
HLA B*0702-transgenic IFN-α/βR−/− mice were bred at the La Jolla Institute for Allergy and Immunology Animal Facility (La Jolla, CA). All mouse experiments were performed following the Institutional Animal Care and Use Committee–approved animal protocols. Age- and sex-matched mice between 5–6 weeks of age and both male and female mice were used in all experiments.
The mouse-adapted dengue virus 2 (DENV2), D2S10, was derived from the clinical isolate PL046. D2S10 was obtained as described (Shresta et al., 2006) by passaging parental virus PL046 through mice and C6/36 mosquito cells 10 times. S221 is a triple-plaque purified clone derived from D2S10 (Zellweger et al., 2013, Zellweger et al., 2014). The same preparation of S221 was used for this entire project. Mice were inoculated with a dose of 1 × 1010 genomic equivalents (GE) S221 for challenge after immunization or 5 × 108 GE for Intracellular Cytokine Staining (ICS) and ELISPOT. For all experiments with DENV3, EDEN3, a clinical isolate derived from the Early Dengue Infection and Outcomes Study (EDEN) (Low et al., 2006) was obtained from Dr. Subhash Vasudevan, and mice were infected with 1 × 1010 GE.
2.2. Peptide Synthesis
All peptides were synthesized by A&A Lab LLC, San Diego. Peptides for ELISPOT were synthesized as crude material and mass spectral analysis of each peptide was performed to validate the synthesis. All 9-mer and 10-mer peptides for flow cytometry and immunization were synthesized and purified by reverse-phase HPLC to ≥ 95% purity. Peptides were stored at − 20 °C after being dissolved in DMSO and aliquoted to avoid freeze-thaw damage.
2.3. Ex Vivo Interferon-gamma (IFN-γ) Enzyme-linked Immunosorbent Spot (ELISPOT) Assay
CD8 T cells were isolated by magnetic bead positive selection (Miltenyi Biotec, Germany). A total of 2 × 105 CD8 T cells were stimulated with 1 × 105 naive splenocytes as antigen presenting cells and 10 μg/ml individual DENV2-specific peptides in 96-well flat-bottom plates (Immobilon-P; Millipore, MA) coated with anti–IFN-γ monoclonal antibody (mAb) (clone AN18; Mabtech, Sweden) in triplicate. Wells were washed with PBS/0.05% Tween 20 after 20 h incubation at 37 °C and then incubated with biotinylated IFN-γ mAb (clone R4-6A2; Mabtech) for 2 h. The spots were developed using Vectastain Elite ABC peroxidase (Vector Laboratories, Burlingame, CA) and 3-amino-9-ethylcarbazole (Sigma-Aldrich, St. Louis, MO) and counted by computer-assisted image analysis (KS-ELISPOT reader; Zeiss, Germany).
2.4. MHC/Peptide Binding
Purification of HLA class I molecules and quantitative competitive inhibition assays to measure the binding affinity of peptides to purified MHC were performed as described elsewhere (Sidney et al., 2013). Briefly, 0.1–1 nM high-affinity radiolabeled peptide was coincubated at room temperature (RT) with 1 μM to 1 nM purified MHC in the presence of protease inhibitors and 1 μM β2-microglobulin. Following 2-days incubation, MHC-bound radioactivity was determined by capturing MHC/peptide complexes on W6/32 (anti-class I) Ab-coated Lumitrac 600 plates (Greiner Bio-One, Germany) and measuring bound cpm using the TopCount microscintillation counter (Packard Instrument Company, CT), and the concentration of peptide yielding 50% inhibition of the binding of the radiolabeled peptide was calculated. Under the conditions used, where [label] < [MHC] and IC50 ≥ [MHC], the measured IC50 values were reasonable approximations of true Kd values. Each competitor peptide was tested at six different concentrations covering a 100,000-fold dose range, and in three or more independent experiments. As a positive control, the unlabeled version of the radiolabeled probe was also tested in each experiment.
2.5. Flow Cytometric Analyses
Mice were sacrificed by isoflurane inhalation and spleens were collected in 10% FBS media. For ICS, splenocytes were counted after red blood cell lysis and were plated as 2 × 106 splenocytes/well in in 96-well U-bottom plates. Cells were stimulated with individual or pooled DENV2 peptides (5 μg/ml of each peptide) or their respective variants for 6 h, in the presence of Brefeldin A (GolgiPlug; BD Biosciences) and anti-CD107a PE (clone 1D4B, eBioscience) during the 4 last hours. Different concentrations of pooled peptides (5, 0.5, 0.05 and 0.005 μg/ml) were tested in T cell avidity assays.
Cells were washed and labeled with anti-CD3 PerCpCy 5.5 (Clone 145-2C11, TONBO), anti-CD8 PE-Cy7 (clone 53-67, BD biosciences), anti-CD44 eFluor 450 (clone IM7, eBioscience), anti-CD62L APC eFluor 780 (clone Mel-14, eBioscience). Then, cells were fixed and permeabilized using the BD Cytofix/Cytoperm Kit, and stained with a combination of anti-IFNγ FITC (clone XMG 1.2, TONBO) and anti-TNFα APC (clone MP6-XT22, eBioscience). Samples were read on an LSR II (BD Biosciences) and were analyzed using FlowJo software X 10.0.7 (Tree Star, Ashland, OR).
2.6. Peptide Immunization
HLA B*0702-transgenic IFN-α/βR−/− mice were immunized subcutaneously (s.c) with a combination of 100 μg of HCV-core helper peptide (I-Ab restricted, TPPAYRRPPNAPIL) and 50 μg of each five DENV2 peptides NS31682–1690, NS31700–1709, NS32070–2078, NS4B2280–2289, NS52885–2894 or their corresponding variant peptides from DENV1/3/4 emulsified in Complete Freund Adjuvant (CFA) (Difco, Detroit, MI). Mock-immunized mice received a combined injection of helper peptide and DMSO. Following 21 days post-priming, mice were boosted with the same mixture as noted above emulsified in Incomplete Freund Adjuvant (IFA) and challenged nine days later with 1 × 1010 GE of DENV2 strain S221 or 1 × 1010 GE of DENV3 strain EDEN3 intravenously (i.v.). Three days post-challenge, organs were harvested after PBS perfusion and blood was collected by cardiac puncture. RNA was isolated, and DENV2 or DENV3 viral RNA levels were measured by real-time qRT-PCR.
2.7. Statistical Analyses
All data were analyzed with Prism software version 5.0 (GraphPad Software, Inc., San Diego, CA) and expressed as mean ± SEM. Statistical significance was determined using unpaired t-test with Welch's correction for the viral RNA levels. Two ways ANOVA or Kruskal Wallis tests were used to compare > 2 groups. p < 0.05 was considered as significant.
3. Results
3.1. Selection of Immunodominant T Cell Response Induced by DENV2 Infection
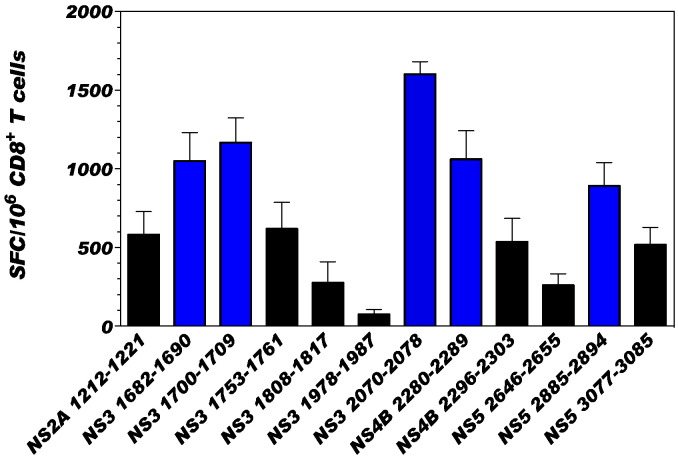
To investigate the immunogenicity and immunodominance of class I restricted CD8 T cells of human relevance, HLA B*0702-transgenic IFN-α/βR−/− mice were used. This particular HLA transgenic mouse model was selected because our study using human PBMC from DENV-exposed blood donors revealed that the HLA B*0702 allele is associated with mild disease and T cell responses high in frequency and magnitude; in contrast, HLA alleles such as A*0101 and A*2401 are associated with severe dengue disease and weak DENV-specific T cell response (Weiskopf et al., 2013). Based on our published study that identified DENV 2 specific epitopes in HLA B*0702-restricted IFN-α/βR−/− mice (Weiskopf et al., 2011), we first evaluated the ability of the 12 DENV2 epitopes identified in that study to induce a strong T cell response in HLA-B*0702-transgenic IFN-α/βR−/− mice. The capacity of each of the 12 DENV2 peptides to induce a T cell response was evaluated by IFNγ ELISPOT (Fig. 1). We selected 5 peptides that elicited high magnitude T cell responses (> 800 SFC/106 CD8 T cells: NS31682–1690 (1050 ± 405 SFC/106), NS31700–1709 (1166 ± 351 SFC/106), NS32070–2078 (1601 ± 179 SFC/106), NS4B2280–2289 (1061.33 ± 404.84 SFC/106) and NS52885–2894 (891.00 ± 331.63 SFC/106)). This immunodominance pattern is in accordance with published studies demonstrating that DENV NS3, NS4B, and NS5 are the main targets of the CD8 T cell response (Duangchinda et al., 2010, Weiskopf et al., 2013).
Fig. 1.
CD8 T cell reactivity against DENV2-specific HLA-B*0702 restricted epitopes.
Previously identified DENV2 specific epitopes were tested in an IFNγ ELISPOT assay to determine response hierarchies after infection with DENV2. ELISPOT was performed using CD8 T cells isolated from HLA-B*0702-transgenic type I IFN receptor knockout C57BL/6 mice (IFN-α/βR−/−) seven days after intravenous infection with 5 × 108 GE of DENV2. CD8 T cells were stimulated with naive splenocytes as antigen presenting cells (APC) and 10 μg/ml of individual peptides. The data are expressed as the mean number of SFC per 106 CD8 T cells. Peptides were tested in triplicates in five independent experiments (total n = 20 mice). Average responses between all experiments are shown and the error bars are presented in SEM. Peptide inducing robust T cell responses (> 800 SFC/106 CD8 T cells) are represented with blue bars.
We compared the amino acid sequence of DENV2 to the sequence at the same position in other serotypes (DENV1/3/4) for each of the 5 immunodominant epitopes. The sequence and the position of each epitope in the polyprotein are summarized in Table 1. DENV2 peptides can be characterized into two groups in terms of their pattern of sequence homology: conserved or DENV2-specific. Three of the 5 DENV2 peptides were conserved—i.e. the same peptide sequence existed in 2 or more serotypes. The other 2 peptides were DENV2-specific—i.e. at least one amino acid of the DENV2 peptide sequence was different in DENV1, DENV3, and DENV4 (DENV1/3/4). We refer to these alternative sequences with at least one amino acid difference from DENV2 as cross-reactive variants. For example, NS31682–1690 is conserved (same amino acid sequence) in DENV2, DENV1, and DENV3, and the corresponding peptide in DENV4 is a cross-reactive variant with a different amino acid in two positions. NS32070–2078 is DENV2-specific, as the DENV2 sequence is different than DENV1, DENV3, and DENV4. The homology seen in conserved peptides is due to the high amino acid conservation between the four DENV serotypes (67–75%) (Kuno et al., 1998). No variation of the T cell response was observed regarding the pattern of sequence homology in DENV2 peptides (specific and conserved peptides).
Table 1.
Characterization of DENV2 peptides and their corresponding variant peptides from other serotypes.
The position, sequence, length, and the serotype of each of five peptides inducing a strong T cell response are represented. The changed amino acid in each variant peptide is indicated in red. The half maximal inhibitory concentration IC50 (nM) defined the binding affinity for the MHC of each peptide. The magnitude of T cell response was assessed using ELISPOT IFNγ in SFC/106 of CD8 T cells. All of the data are expressed in standard error of mean (SEM). The percentage of IFNγ production upon stimulation by variant peptides in comparison to DENV2 peptides are represented in blue.
3.2. The Magnitude of the Cross-reactive CD8 T Cell Response is Either Equal or Lower Magnitude Than DENV2 Serotype-specific T Cell Response
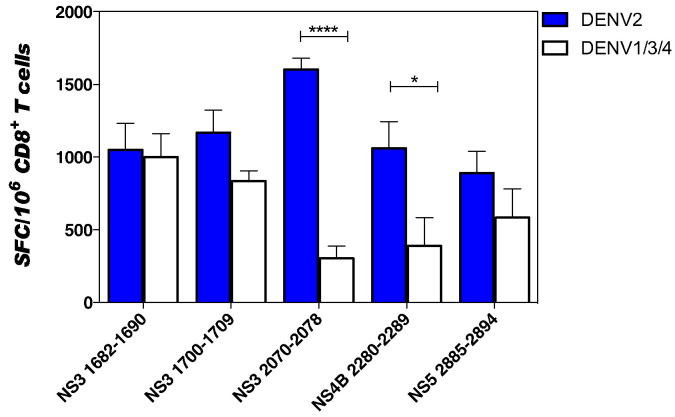
We next compared the CD8 T cell response induced by variant peptides relative to the 5 immunodominant epitopes in an IFNγ ELISPOT analysis (Fig. 2 and Table 1). The DENV2-exposed T cell responses varied in response to variant peptides (Fig. 2). The T cell response to variant peptide sequence ranged from equal (96.5% of the DENV2 magnitude for the DENV4 NS31682–1690) to low responses with only 33.19% of the DENV2 magnitude for DENV4 epitope NS32070–2078 (Table 1). These results indicate the presence of cross-reactive T cells after DENV2 infection.
Fig. 2.
CD8 T cells response after stimulation by DENV2-specific or DENV1/3/4 variant peptides.
IFNγ ELISPOT was performed using CD8 T cells isolated from HLA-B*0702-transgenic IFN-α/βR−/− seven days after intravenous infection with 5 × 108 GE of DENV2. CD8 T cells were stimulated with naive splenocytes as APCs and 10 μg/ml of individual peptide. All five DENV2-specific peptides that induced strong T cell responses are shown in blue (DENV2) and their corresponding variant peptides from other serotypes in white (DENV1/3/4). The data are expressed as the mean number of SFC per 106 CD8 T cells and correspond to an average of five independent experiments performed in triplicate. The error bars are presented in SEM. Two way ANOVA, multiple comparisons was used for statistical test, * < 0.05 and ****p < 0.0001.
In terms of cross-reactivity, two variant peptides, NS32070–2078 and NS4B2280–2289 with 33.19 and 40% of CD8 T cell response, respectively, elicited notably lower magnitude T cell responses compared to their corresponding DENV2 sequence. Both the number (1, 2, or 3) and position (first, last, or middle position in peptide) of the amino acid changes did not seem to influence the magnitude of the CD8 T cell response. For instance, the cross-reactive variant peptide that best matched the DENV2 peptide in terms of the magnitude of the T cell response (NS31682–1690) differs in two amino acids, whereas the NS4B2280–2289 epitope in DENV1 or DENV3 that differs in only one amino acid from DENV2 elicited 40% of DENV2-specific T cell response.
To determine if binding efficiency could affect the T cell response, we performed HLA binding assays with each peptide (specific, conserved, or variant, Table 1) as previously described (Sidney et al., 2013). All peptides showed a high affinity to the MHC molecule, as indicated by an IC50 ≤ 10 nM independent of the number of amino acid changes. Thus, binding efficiency is not likely responsible for the significant reduction in some of the cross-reactive T cell responses. In particular, the binding affinities of the NS32070–2078 and NS4B2280–2289 variant epitopes were 1.77 and 6.15 nM, respectively, which were equal to or comparable to the value of their DENV2 counterparts, albeit the cross-reactive and DENV2-specific T cell magnitudes were different.
3.3. The Cross-reactive CD8 T Cell Response Is Multifunctional in Terms of Cytokine Production
To assess the multi-functionality of the cross-reactive vs. DENV2-specific CD8 T cells, we performed intracellular cytokine staining (ICS). Splenocytes were isolated from HLA-B*0702-transgenic IFN-α/βR−/− mice infected with 5 × 108 GE of DENV2 (S221 strain) and stimulated ex vivo with single or pooled peptides (Corresponding to a mix of all five immunodominant epitopes for DENV2 or all five peptides used as variant in other serotypes) in the presence of Brefeldin A (BFA) and CD107a for 4 h. Cells were stained for intracellular cytokines IFNγ and TNFα. All infected mice had twice as many CD8 T cells in spleens compared to uninfected control animals (data not shown), indicating an efficient infection.
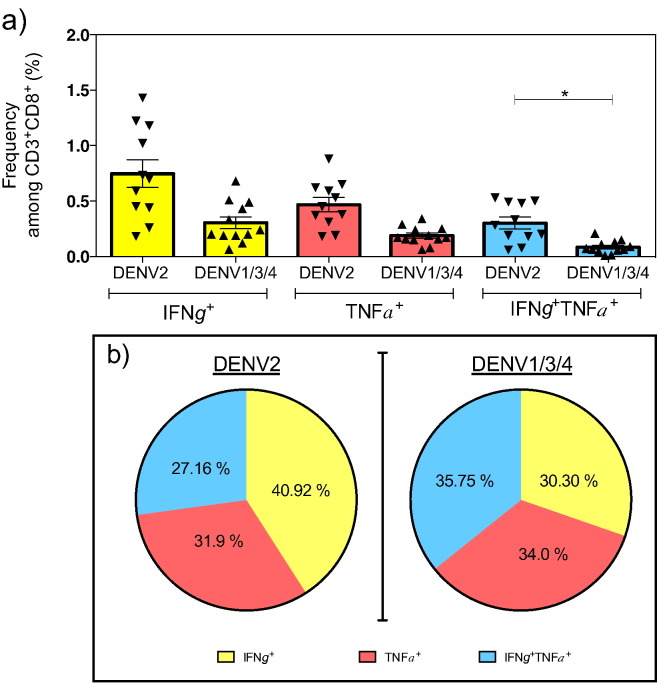
The percentage of cells producing IFNγ and TNFα upon stimulation with DENV2 specific peptides or their corresponding variants from DENV1/3/4 is represented in Fig. 3a. The frequencies of the single positive, DENV2-exposed CD8 T cells were higher after stimulation with the DENV2 peptide pool (0.74 ± 0.12% and 0.46 ± 0.06%, respectively, for IFNγ and TNFα) than the variant peptide pool (0.30 ± 0.05% and 0.18 ± 0.02%, respectively, for IFNγ and TNFα), but this difference was not significant. However, the percentage of double positive cells (IFNγ+ TNFα+) in response to the DENV2 peptide pool (0.30 ± 0.05%) was significantly higher than to variant peptide pool (0.08 ± 0.01%), revealing that the DENV2-exposed CD8+ T cell population contained a significantly higher frequency of IFNγ+ TNFα+ double positive cells following stimulation with pooled DENV2 peptides than cross-reactive peptides (Fig. 3a).
Fig. 3.
Functional phenotype of serotype-specific vs. cross-reactive CD8 T cells.
Mice were infected with 5 × 108 GE of DENV2 (Infected) or injected with 10% FBS-PBS (naïve). Seven days later, spleens were harvested and ICS was performed with individual or previously selected pooled peptides. (a) The frequency of CD8 T cells expressing IFNγ (Yellow), TNFα (Salmon) or IFNγTNFα (Blue) in infected mice without the background produced by naïve mice. (b) The cytokine profile of CD8 T cells in response to DENV2 or DENV1/3/4 peptide stimulation, as illustrated in pie-charts. These pie-charts show the percentage of IFNγ (Yellow), TNFα (Salmon) or IFNγTNFα (Blue) frequencies compared to whole cytokine production. (c) The cytokine profile induced by stimulation with individual peptide. (d) The frequency of double positive cells for IFNγ and CD107a produced by splenocytes from infected (Black bar) or naïve (White bar) mice.
Experiments were performed twice with a total number of 11 infected mice for DENV2 stimulation, 12 infected mice for DENV1/3/4 stimulation, and 9 naïve mice. Each independent experiment showed similar results. The error bars are represented in SEM. Kruskal-Wallis multiple comparisons was used for statistical test. * < 0.05.
In terms of the cytokine profile, DENV2-exposed CD8 T cells elicited a similar pattern of cytokines in the presence of DENV2 or DENV1/3/4 peptides (Fig. 3b). The pie-chart is divided into three parts showing that the DENV2-exposed CD8 T cell population included 40.9% of IFNγ+ cells, 31.9% of TNFα+ cells, and 27.6% of double positive cells after stimulation with the DENV2 peptide pool, and 30.3% of IFNγ+ cells, 34.0% of TNFα+ cells, and 35.8% of double positive cells after stimulation with the variant peptide pool (Fig. 3b). Further analysis using individual peptide instead of peptide pools revealed that the cytokine profile of DENV2-exposed CD8 T cells upon stimulation with each individual DENV2 peptide presented the same pattern represented by an equal distribution of IFNγ+, TNFα+, and IFNγ+ TNFα+ cells (Fig. 3c, left panel). A similar pattern has been identified for three of the five individual variant peptides (Fig. 3c right panel, NS31682–1690, NS31700–1709, NS52885–2894 epitopes). Two of the individual variant peptides seem to have a different pattern in favor of IFNγ production: NS32070–2078 and NS4B2280–2289 with 67.57% and 96.49%, respectively (Fig. 3c). However, T cells stimulated by pooled peptides (DENV2 or DENV1/3/4) reveal the same pattern. Over all, these results show that no specific cytokine pattern was observed in DENV1/3/4 cross-reactive vs. DENV2 serotype-specific CD8 T cell responses.
CD107a is present on the inner membrane of vesicles containing perforin and granzymes, so positive staining defines degranulation of activated T cells (Alter et al., 2004). In all our experiments, > 95% of IFNγ producing cells are CD107a positive cells (data not shown). CD107a, combined with IFNγ, determine CD8 T cells capable of eliminating virus-infected cells (Fig. 3d). DENV2 peptides induced a higher percentage of CD107a+ IFNγ+ CD8 T cells (0.27 ± 0.09%) compared to DENV1/3/4 peptides (0.06 ± 0.01%).
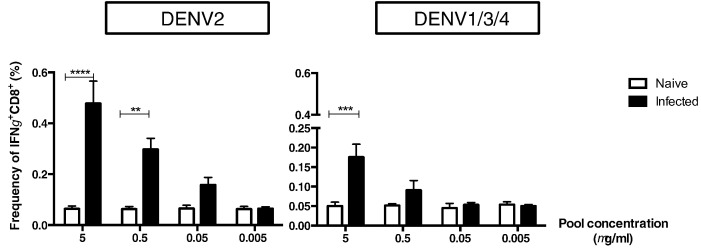
Based on studies suggesting that cross-reactive T cells with low affinity are activated during secondary DENV infection in humans (Mongkolsapaya et al., 2003, Rothman et al., 2014, Screaton et al., 2015), we next determined the functional avidity of DENV2-exposed CD8 T cells in response to cross-reactive peptides. ICS was performed to measure IFNγ production by DENV2-exposed T cells upon stimulation with different concentrations of DENV2 or variant peptide pool. A response was considered positive if IFNγ production was significantly higher than measured in background experiments (Fig. 4. naïve mice, white bars). The DENV2 peptide pool was able to induce production of IFNγ at lower concentrations (0.5 and 0.05 μg/ml, only significantly higher for 0.5 μg/ml) than the variant peptide pool (Fig. 4), indicating that DENV2-exposed T cells have a lower avidity response upon stimulation with cross-reactive peptides than with DENV2 peptides.
Fig. 4.
Avidity of serotype-specific vs. cross-reactive CD8 T cells.
Mice were infected with 5 × 108 GE of DENV2 (black bar) or injected with 10% FBS-PBS (white bar). Seven days later, spleens were harvested and ICS was performed with different concentrations (5, 0.5, 0.05, or 0.005 μg/ml) of pooled peptides from DENV2 or other serotypes DENV1/3/4. The frequency of IFNγ was evaluated at each concentration and for both conditions. Two independent experiments were performed with 6 mice per group. Each experiment showed similar trend. All data are expressed in standard error of mean (SEM). Kruskal-Wallis multiple comparisons was used for statistical test, ** < 0.005; ***p < 0.001; ****p < 0.0001.
3.4. Immunization With DENV1/3/4 Variant Peptides Can Protect Against DENV2 or DENV3 Infection in HLA-B*0702-Transgenic IFN-α/βR−/− Mice
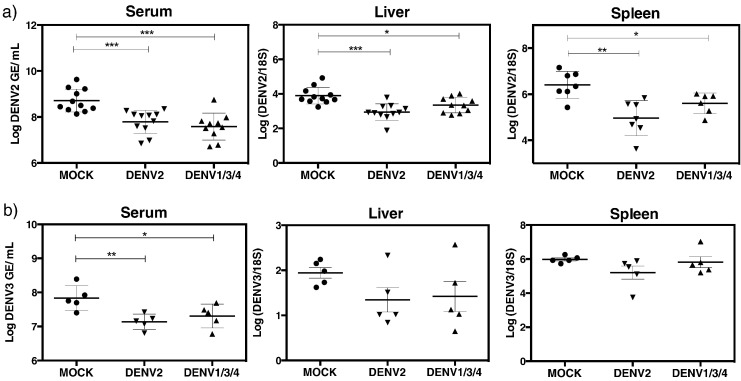
To investigate whether variant peptides can contribute to protection against DENV infection, our previously published vaccination strategy was adopted (Yauch et al., 2009, Yauch et al., 2010). One group of HLA-B*0702-transgenic IFN-α/βR−/− transgenic mice received an injection (s.c.) of the five DENV2 peptide cocktail in Complete Freund Adjuvant (CFA), while the other experimental group was administered the corresponding variant peptide cocktail. A control group was included in which mice were treated with only the helper peptide and DMSO. Twenty-one days after the first immunization, mice were boosted with the same peptides in Incomplete Freund Adjuvant (IFA). Nine days after the boost, mice were challenged with 1 × 1010 GE of DENV2 (S221 strain), followed by quantification of DENV2 RNA levels in tissues by qRT-PCR on day 3 after challenge. Mice immunized with DENV2 peptides or DENV1/3/4 variant peptides had significantly lower levels of DENV2 RNA in serum, spleen, and liver as compared to the mock-immunized group (Fig. 5a). These results imply that DENV1/3/4-cross-reactive T cells can contribute to protection against DENV2 infection. To validate and extend these results to protection against another DENV serotype, we used the same immunization protocol described above and challenged mice with DENV3. DENV3 RNA levels in the serum of DENV2- or DENV1/3/4-immunized mice were lower as compared to mock-immunized mice (Fig. 5b). DENV3 RNA levels in the liver and spleen of the peptide-immunized groups trended to decrease relative to mock-immunized mice, although no significant difference was observed between peptide-immunized and mock-immunized animals. Taken together, these results demonstrate that DENV serotype-cross-reactive CD8 T cells can mediate protection.
Fig. 5.
DENV2 and DENV3 viral burden in peptide-immunized mice.
Mice were immunized with a mix of five DENV2 peptides selected (DENV2) or their corresponding variants (DENV1/3/4), followed by boosting with the same mixture 21 days later. Nine days post-boosting, mice were challenged with DENV at 1 × 1010 GE and DENV RNA levels were quantified by real time qRT-PCR, three days post-challenge. (a) DENV2 and (b) DENV3 RNA levels in serum, liver, and spleen are represented in log scale. The results were pooled from three independent experiments for DENV2. Unpaired t-test with Welch's correction was used as a statistical test, * < 0.05 was defined as significant, and all error bars are represented in SEM.
3.5. Cross-reactive T Cells Elicit a Polyfunctional T Cell Response In Vivo
To confirm that cross-reactive T cells were responsible for protection against DENV2 challenge in the peptide-immunized mice, we evaluated the CD8 T cell response in the immunized mice via ICS just prior to and 3 days after viral challenge. CD44 and CD62L expression was used to discriminate the effector memory subset of CD8 T cells, and antigen-specific cells were assessed based on IFNγ expression by CD44+ CD62L− effector memory subset (Fig. 6a).
Fig. 6.
CD8 T cell response in mice immunized with DENV2 or DENV1/3/4 variant peptides before and after DENV2 challenge.
Mice were immunized with a mix of five DENV2 peptides (DENV2) or their corresponding variants (DENV1/3/4) and were boosted with the same mixture 21 days later. Nine days after boosting, spleens were harvested and splenocytes were stimulated with either individual or pooled DENV2 peptides in presence of Brefeldin A and CD107a. (a) The gating strategy to select effector memory (EM) or central memory (CM) IFNγ positive CD8 T cells is illustrated. (b) The frequencies of effector memory CD8 T cells producing IFNγ 30 days after immunization (left panel) and the frequencies obtained three days after challenge with 1 × 1010 GE DENV2 (right panel) are shown. (c) The frequencies of central memory cells producing IFNγ in all three groups before and after challenge with DENV2 are represented. (d) CD107a and IFNγ double expression in mock (white), DENV2 (black), and DENV1/3/4 (grey) peptide-immunized mice before (left panel) and after challenge (right panel) is shown. The results were pooled from two independent experiments with n = 7 mice/group before challenge and n = 8 mice per group after challenge. All data are expressed in standard error of mean (SEM). Kruskal-Wallis multiple comparisons was used as a statistical test. * < 0.05, ** < 0.005.
Just prior to viral challenge on day 9 after the boost, mice immunized with DENV2 peptides had a higher percentage of IFNγ+ effector memory CD8 T cells (0.78 ± 0.11%) than mice that were mock-immunized (0.19 ± 0.07%) or immunized with variant peptides (0.23 ± 0.05%) upon stimulation with DENV2 peptides (Fig. 6b, left panel). In contrast, no difference was observed in the frequency of IFNγ-producing central memory subsets (identified based the expression of CD44 and CD62L) in peptide-immunized vs. mock-immunized mice. Approximately 0.2 ± 0.04%, 0.21 ± 0.05%, and 0.15 ± 0.04%, respectively, were IFNγ-producing central memory CD8 T cells in the mock-, DENV2 peptide-, and DENV1/3/4 peptide-immunized mice before challenge (Fig. 6c, left panel). After challenge, the frequency of IFNγ-producing central memory CD8 T cells was similar to that obtained before challenge (Fig. 6c, right panel).
Mice immunized with DENV2 peptides also tended to have higher frequency of potentially cytotoxic (i.e. CD107a+ IFNγ+) CD8 T cells than the other two groups of animals, albeit this difference was not significant (Fig. 6d, left panel). Three days after DENV2 challenge of the immunized mice, mice immunized with DENV2 peptides or variant peptides contained significantly higher frequency of IFNγ+ effector memory CD8 T cells upon stimulation with DENV2 peptides (Fig. 6b, right panel). Both groups of peptide-immunized mice also seemed to contain higher percentages of potentially cytotoxic CD107a+ IFNγ+ CD8 T cells relative to mock-immunized mice (Fig. 6d, left panel). Taken together, results shown in Fig. 5, Fig. 6 demonstrate the presence of multifunctional DENV serotype-cross-reactive T cells that can mediate viral clearance.
4. Discussion
Epidemiological studies have highlighted an increase of severe disease manifestations (DHF/DSS) during secondary infections with different DENV serotype (Halstead, 2007). To explain this epidemiological observation, two dominant hypotheses have been postulated: ADE and T cell original antigenic sin. The relevance of ADE to DHF/DSS pathogenesis had been controversial until the demonstration of ADE in vivo in 2010, when our laboratory and another group independently demonstrated that subneutralizing levels of antibody can turn a mild illness into a lethal disease in mice lacking IFN-α/β receptor alone or both IFN-α/β and γ receptors (Zellweger et al., 2010, Balsitis et al., 2010). Unlike wild-type mice, the IFN receptor-deficient mice support high levels of DENV replication and in the ADE model of infection, develop signs of disease that mimic DHF/DSS in humans (Zellweger et al., 2010). In contrast, no study to date has provided direct evidence in support of the original T cell antigenic sin hypothesis. As a step towards addressing this hypothesis, in this study, we investigated the contribution of DENV serotype-specific vs. cross-reactive CD8 T cells to protection against DENV infection using an HLA transgenic mouse model of human relevance. Our results demonstrated that both serotype-specific and cross-reactive CD8 T cells could reduce DENV burden in tissues, providing no direct evidence for the original T cell antigenic sin hypothesis. Instead, our findings support recent data implicating HLA-linked protective role for CD8 T cells against DENV infection in humans (Weiskopf et al., 2013).
HLA-transgenic mice are widely used to model the human T cell response in mice. We focused the present study on the HLA B*0702 allele, which is associated with strong T cell response in terms of magnitude and breath (Weiskopf et al., 2013), thereby allowing us to test the role of multiple cross-reactive epitopes using our HLA B*0702-transgenic IFN-α/βR−/− mouse model of DENV2 infection. Studies with DENV-exposed human PBMC have shown that serotype cross-reactive T cells can be preferentially activated and can exhibit an altered phenotype in terms of cytokine production, degranulation, and avidity (Mangada and Rothman, 2005, Mongkolsapaya et al., 2003, Mongkolsapaya et al., 2006). In particular, Rothman and colleagues reported a lower affinity of CD8 T cells for the sequence from another serotype during secondary infection and preferential cytokine production in favor of TNFα by cross-reactive CD8 T cells (Bashyam et al., 2006). Screaton and colleagues similarly reported activation of cross-reactive CD8 T cells with low avidity in DHF/DSS patients (Mongkolsapaya et al., 2003). Based on these findings, cross-reactive T cells have been hypothesized to play a pathogenic role during DENV infection. Our results, however, show that cross-reactive T cells do not appear to exhibit an altered cytokine profile and that cross-reactive T cells can decrease viral burden in tissues despite having lower magnitude and avidity than serotype-specific T cells. Our findings are consistent with an increasing number of new studies implicating a protective role for T cells during DENV infection in humans. In addition to our studies revealing an association between HLA-linked T cell responses and disease susceptibility (Weiskopf et al., 2013, Weiskopf et al., 2015b), an earlier study by a different group observed that the breadth and magnitude of the CD8 T cell response during secondary DENV infection was not significantly associated with disease severity (Simmons et al., 2005). Most recently, another group reported that the expansion of effector memory CD8 T cells during acute DENV infection correlated with reduced viremia (de Matos et al., 2015).
Using H-2b mice (i.e. non HLA-transgenic) lacking IFN-α/β receptor alone or both IFN-α/β and γ receptors, we have previously demonstrated that CD8 T cells can play a critical role in protection against DENV in the context of primary infection (Yauch et al., 2009, Prestwood et al., 2012), vaccination (Zellweger et al., 2013), heterotypic secondary infection (Zellweger et al., 2015), and ADE (Zellweger et al., 2014). The present study with HLA transgenic mice highlights the role of serotype-specific and cross-reactive CD8 T cells in protection against DENV infection in the context of epitopes that are recognized by humans. Collectively, our studies with mouse models and new studies supporting a protective role for CD8 T cells against DENV infection in individuals from endemic countries imply that a dengue vaccine should elicit a robust, long-lasting CD8 T cell response. Based on the present results, this optimal CD8 T cell response should ideally include a combination of each of the four DENV serotype-specific, conserved (i.e. identical in more than one serotype), and cross-reactive CD8 T cells that are polyfunctional.
However based on the present study, we cannot reject the possibility that cross-reactive T cells are pathogenic at late stages of DHF/DSS development. For instance, pathogenic T cells may be a consequence of DHF/DSS rather than a cause. In this scenario, cross-reactive T cells are protective in most secondary infection cases but some unknown factors induces an irregularity in the environment that switch the function of T cells from protective to pathogenic in DHF/DSS patients. Studies investigating the role of cross-reactive CD8 T cells in the context of severe vs. mild disease using IFN-α/βR−/− mice that are transgenic for particular alleles associated with strong vs. weak T cell responses may reveal the potentially pathogenic role of cross-reactive T cells during DENV infection.
Consistent with our overall assertion that serotype-specific, conserved, and cross-reactive T cells all can contribute to protection against DENV infection, phase 2b and 3 trials of Sanofi's DENV-yellow fever virus (YFV) chimeric vaccine showed limited efficacy depending on the serotype and host immune status (Guy et al., 2015, Guy and Jackson, 2016, Hadinegoro et al., 2015, Halstead and Russell, 2016). In particular, the Sanofi vaccine contains YFV but not DENV NS proteins, which are the major targets of the anti-DENV CD8 T cell response (Weiskopf et al., 2013), suggesting that the absence of the vaccine-induced DENV-specific CD8 T cell response may be responsible for the lack of robust protection observed in the Sanofi clinical trials. In comparison with the Sanofi vaccine, the other two leading vaccine candidates developed by NIH and Takeda are also live-attenuated viruses, but are not chimeric of two different flaviviruses. The NIH and Takeda vaccine candidates represent only DENV, and thus contain DENV NS proteins. However, the NIH tetravalent vaccine formulation contains NS proteins from 3 DENV serotypes, while the Takeda tetravalent formulation consists of NS proteins from only 1 DENV serotype. Phase 1 trials have shown that both the NIH and Takeda vaccine can induce DENV-specific T cell responses (Weiskopf et al., 2014, Chu et al., 2015). While the NIH vaccine-induced CD8 T response in phase 1 participants focused on conserved epitopes (i.e. identical in > 1 serotype) (Weiskopf et al., 2014), the specificity of the Takeda vaccine-induced CD8 T cell response is as yet to be fully defined and may differ from the NIH vaccine by targeting DENV2 serotype-specific, conserved, and cross-reactive epitopes. Our finding that cross-reactive T cells with even low avidity and magnitude can limit viral burden in mice imply that both NIH and Takeda vaccines may show good efficacy in phase 2 and 3 trials. In summary, our results emphasize that a DENV vaccine should elicit not only Ab but also CD8 T cell responses to maximize efficacy.
5. Conclusion
The T cell response to DENV has been implicated to play a dual role in both protection and pathogenesis. In particular, serotype-cross-reactive T cells have been hypothesized to contribute to DENV pathogenesis. However, no study to date has provided direct evidence supporting a pathogenic role for T cells during DENV infection. To address the potentially pathogenic role of DENV antigen-specific T cells, the present study evaluated the role of serotype-specific vs. cross-reactive CD8 T cells with known antigen specificity using a HLA transgenic mouse model of DENV infection and a peptide immunization setting. The results show that both serotype-specific and cross-reactive CD8 T cells can contribute to protection against DENV challenge, consistent with emerging literature implicating an important role for T cells in protection against DENV infection.
Conflict of Interest Statement
All authors declare no conflict of interest.
Author Contributions
Annie EN: study design, data collection, analysis, interpretation, writing, literature search.
Hui-Wen C: study, data collection and analysis, writing.
William T: data collection and analysis.
Yunichel J: data collection and analysis.
Kevin K: data collection and analysis.
Daniela W: data interpretation.
John S: data collection, data interpretation.
Alessandro S: data interpretation.
Sujan S: study design, data interpretation, writing.
Acknowledgements
We thank Dr. Kenneth Kim for critical reading of the manuscript.
This project was supported by NIH grants U54AI057517, R56 A1085063, U01 AI082185 and R01 AI116813 to SS and by HHSN27220140045C to AS.
References
- Aguirre S., Maestre A.M., Pagni S., Patel J.R., Savage T., Gutman D., Maringer K. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 2012;8(10) doi: 10.1371/journal.ppat.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter G., Malenfant J.M., Altfeld M. CD107a as a functional marker for the identification of natural killer cell activity. J. Immunol. Methods. 2004;294(1–2):15–22. doi: 10.1016/j.jim.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Ashour J., Morrison J., Laurent-Rolle M., Belicha-Villanueva A., Plumlee C.R., Bernal-Rubio D., Williams K.L. Mouse STAT2 restricts early dengue virus replication. Cell Host Microbe. 2010;8(5):410–421. doi: 10.1016/j.chom.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsitis S.J., Williams K.L., Lachica R., Flores D., Kyle J.L., Mehlhop E., Johnson S., Diamond M.S., Robert Beatty P., Harris E. Lethal antibody enhancement of dengue disease in mice is prevented by fc modification. PLoS Pathog. 2010;6(2) doi: 10.1371/journal.ppat.1000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashyam H.S., Green S., Rothman A.L. Dengue virus-reactive CD8+ T cells display quantitative and qualitative differences in their response to variant epitopes of heterologous viral serotypes. J. Immunol. (Baltimore, Md.: 1950) 2006;176(5):2817–2824. doi: 10.4049/jimmunol.176.5.2817. [DOI] [PubMed] [Google Scholar]
- Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L., Drake J.M. The global distribution and burden of dengue. Nature. 2013;496(7446):504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., George S.L., Stinchcomb D.T., Osorio J.E., Partidos C.D. CD8+ T-cell responses in Flavivirus-naive individuals following immunization with a live-attenuated tetravalent dengue vaccine candidate. J. Infect. Dis. 2015;212(10):1618–1628. doi: 10.1093/infdis/jiv258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Matos A.M., Carvalho K.I., Rosa D.S., Villas-Boas L.S., da Silva W.C., de Lima Rodrigues C.L., Oliveira O.M.N.P.F. CD8+ T lymphocyte expansion, proliferation and activation in dengue fever. PLoS Negl. Trop. Dis. 2015;9(2) doi: 10.1371/journal.pntd.0003520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duangchinda T., Dejnirattisai W., Vasanawathana S., Limpitikul W., Tangthawornchaikul N., Malasit P., Mongkolsapaya J., Screaton G. Immunodominant T-cell responses to dengue virus NS3 are associated with DHF. Proc. Natl. Acad. Sci. U. S. A. 2010;107(39):16922–16927. doi: 10.1073/pnas.1010867107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy B., Jackson N. Dengue vaccine: hypotheses to understand CYD-TDV-induced protection. Nat. Rev. Microbiol. 2016;14(1):45–54. doi: 10.1038/nrmicro.2015.2. [DOI] [PubMed] [Google Scholar]
- Guy B., Briand O., Lang J., Saville M., Jackson N. Development of the Sanofi Pasteur tetravalent dengue vaccine: one more step forward. Vaccine. 2015;33(50):7100–7111. doi: 10.1016/j.vaccine.2015.09.108. [DOI] [PubMed] [Google Scholar]
- Guzman M.G., Halstead S.B., Artsob H., Buchy P., Farrar J., Gubler D.J., Hunsperger E. Dengue: a continuing global threat. Nat. Rev. Microbiol. 2010;8(12 Suppl):S7–16. doi: 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadinegoro S.R., Arredondo-García J.L., Capeding M.R., Deseda C., Chotpitayasunondh T., Reynaldo D., Ismail H.I.H.M. Efficacy and long-term safety of a dengue vaccine in regions of endemic disease. N. Engl. J. Med. 2015;373(13):1195–1206. doi: 10.1056/NEJMoa1506223. [DOI] [PubMed] [Google Scholar]
- Halstead S.B. Dengue. Lancet (London, England) 2007;370(9599):1644–1652. doi: 10.1016/S0140-6736(07)61687-0. [DOI] [PubMed] [Google Scholar]
- Halstead S.B. Controversies in dengue pathogenesis. Paediatr. Int. Child Health. 2012;32(Suppl 1):5–9. doi: 10.1179/2046904712Z.00000000045. (May) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead S.B., Russell P.K. Protective and immunological behavior of chimeric yellow fever dengue vaccine. Vaccine. 2016 doi: 10.1016/j.vaccine.2016.02.004. (February) [DOI] [PubMed] [Google Scholar]
- Jayaratne S.D., Atukorale V., Gomes L., Chang T., Wijesinghe T., Fernando S., Ogg G.S., Malavige G.N. Evaluation of the WHO revised criteria for classification of clinical disease severity in acute adult dengue infection. BMC Res. Notes. 2012;5:645. doi: 10.1186/1756-0500-5-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno G., Chang G.J., Tsuchiya K.R., Karabatsos N., Cropp C.B. Phylogeny of the genus Flavivirus. J. Virol. 1998;72(1):73–83. doi: 10.1128/jvi.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low J.G.H., Ooi E.-E., Tolfvenstam T., Leo Y.-S., Hibberd M.L., Ng L.-C., Lai Y.-L. Early dengue infection and outcome study (EDEN) — study design and preliminary findings. Ann. Acad. Med. Singap. 2006;35(11):783–789. [PubMed] [Google Scholar]
- Malavige G.N., Ogg G. Pathogenesis of severe dengue infection. Ceylon Med. J. 2012;57(3):97–100. doi: 10.4038/cmj.v57i3.4701. [DOI] [PubMed] [Google Scholar]
- Mangada M.M., Rothman A.L. Altered cytokine responses of dengue-specific CD4+ T cells to heterologous serotypes. J. Immunol. (Baltimore, Md.: 1950) 2005;175(4):2676–2683. doi: 10.4049/jimmunol.175.4.2676. [DOI] [PubMed] [Google Scholar]
- Mongkolsapaya J., Dejnirattisai W., Xu X.-n., Vasanawathana S., Tangthawornchaikul N., Chairunsri A., Sawasdivorn S. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat. Med. 2003;9(7):921–927. doi: 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- Mongkolsapaya J., Duangchinda T., Dejnirattisai W., Vasanawathana S., Avirutnan P., Jairungsri A., Khemnu N. T cell responses in dengue hemorrhagic fever: are cross-reactive T cells suboptimal? J. Immunol. (Baltimore, Md.: 1950) 2006;176(6):3821–3829. doi: 10.4049/jimmunol.176.6.3821. [DOI] [PubMed] [Google Scholar]
- Perry S.T., Prestwood T.R., Lada S.M., Benedict C.A., Shresta S. Cardif-mediated signaling controls the initial innate response to dengue virus in vivo. J. Virol. 2009;83(16):8276–8281. doi: 10.1128/JVI.00365-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestwood T.R., Morar M.M., Zellweger R.M., Miller R., May M.M., Yauch L.E., Lada S.M., Shresta S. Gamma interferon (IFN-γ) receptor restricts systemic dengue virus replication and prevents paralysis in IFN-α/β receptor-deficient mice. J. Virol. 2012;86(23):12561–12570. doi: 10.1128/JVI.06743-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman A.L., Medin C.L., Friberg H., Currier J.R. Immunopathogenesis versus protection in dengue virus infections. Curr. Trop. Med. Rep. 2014;1(1):13–20. doi: 10.1007/s40475-013-0009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screaton G., Mongkolsapaya J., Yacoub S., Roberts C. New insights into the immunopathology and control of dengue virus infection. Nat. Rev. Immunol. 2015;15(12):745–759. doi: 10.1038/nri3916. [DOI] [PubMed] [Google Scholar]
- Shresta S., Sharar K.L., Prigozhin D.M., Robert Beatty P., Harris E. Murine model for dengue virus-induced lethal disease with increased vascular permeability. J. Virol. 2006;80(20):10208–10217. doi: 10.1128/JVI.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J., Southwood S., Moore C., Oseroff C., Pinilla C., Grey H.M., Sette A. Measurement of MHC/peptide interactions by gel filtration or monoclonal antibody capture. In: Coligan J.E., editor. Current Protocols in Immunology. 2013. Chapter 18 (February): Unit 18.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons C.P., Dong T., Chau N.V., Dung N.T.P., Chau T.N.B., Thao L.T.T., Dung N.T., Hien T.T., Rowland-Jones S., Farrar J. Early T-cell responses to dengue virus epitopes in Vietnamese adults with secondary dengue virus infections. J. Virol. 2005;79(9):5665–5675. doi: 10.1128/JVI.79.9.5665-5675.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D., Yauch L.E., Angelo M.A., John D.V., Greenbaum J.A., Sidney J., Kolla R.V. Insights into HLA-restricted T cell responses in a novel mouse model of dengue virus infection point toward new implications for vaccine design. J. Immunol. (Baltimore, Md.: 1950) 2011;187(8):4268–4279. doi: 10.4049/jimmunol.1101970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D., Angelo M.A., de Azeredo E.L., Sidney J., Greenbaum J.A., Fernando A.N., Broadwater A. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc. Natl. Acad. Sci. U. S. A. 2013;110(22):E2046–E2053. doi: 10.1073/pnas.1305227110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D., Angelo M.A., Sidney J., Peters B., Shresta S., Sette A. Immunodominance changes as a function of the infecting dengue virus serotype and primary versus secondary infection. J. Virol. 2014;88(19):11383–11394. doi: 10.1128/JVI.01108-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D., Bangs D.J., Sidney J., Kolla R.V., De Silva A.D., de Silva A.M., Crotty S., Peters B., Sette A. Dengue virus infection elicits highly polarized CX3CR1+ cytotoxic CD4+ T cells associated with protective immunity. Proc. Natl. Acad. Sci. U. S. A. 2015;112(31):E4256–E4263. doi: 10.1073/pnas.1505956112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D., Cerpas C., Angelo M.A., Bangs D.J., Sidney J., Paul S., Peters B. Human CD8+ T-cell responses against the 4 dengue virus serotypes are associated with distinct patterns of protein targets. J. Infect. Dis. 2015;212(11):1743–1751. doi: 10.1093/infdis/jiv289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D., Angelo M.A., Bangs D.J., Sidney J., Paul S., Peters B., de Silva A.D. The human CD8+ T cell responses induced by a live attenuated tetravalent dengue vaccine are directed against highly conserved epitopes. J. Virol. 2015;89(1):120–128. doi: 10.1128/JVI.02129-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch L.E., Zellweger R.M., Kotturi M.F., Qutubuddin A., Sidney J., Peters B., Prestwood T.R., Sette A., Shresta S. A protective role for dengue virus-specific CD8+ T cells. J. Immunol. (Baltimore, Md.: 1950) 2009;182(8):4865–4873. doi: 10.4049/jimmunol.0801974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauch L.E., Prestwood T.R., May M.M., Morar M.M., Zellweger R.M., Peters B., Sette A., Shresta S. CD4+ T cells are not required for the induction of dengue virus-specific CD8+ T cell or antibody responses but contribute to protection after vaccination. J. Immunol. (Baltimore, Md.: 1950) 2010;185(9):5405–5416. doi: 10.4049/jimmunol.1001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.-Y., Chang T.-H., Liang J.-J., Chiang R.-L., Lee Y.-L., Liao C.-L., Lin Y.-L. Dengue virus targets the adaptor protein MITA to subvert host innate immunity. PLoS Pathog. 2012;8(6) doi: 10.1371/journal.ppat.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellweger R.M., Prestwood T.R., Shresta S. Enhanced infection of liver sinusoidal endothelial cells in a mouse model of antibody-induced severe dengue disease. Cell Host Microbe. 2010;7(2):128–139. doi: 10.1016/j.chom.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellweger R.M., Miller R., Eddy W.E., White L.J., Johnston R.E., Shresta S. Role of humoral versus cellular responses induced by a protective dengue vaccine candidate. PLoS Pathog. 2013;9(10) doi: 10.1371/journal.ppat.1003723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellweger R.M., Eddy W.E., Tang W.W., Miller R., Shresta S. CD8+ T cells prevent antigen-induced antibody-dependent enhancement of dengue disease in mice. J. Immunol. (Baltimore, Md.: 1950) 2014;193(8):4117–4124. doi: 10.4049/jimmunol.1401597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellweger R.M., Tang W.W., Eddy W.E., King K., Sanchez M.C., Shresta S. CD8+ T cells can mediate short-term protection against heterotypic dengue virus reinfection in mice. J. Virol. 2015;89(12):6494–6505. doi: 10.1128/JVI.00036-15. [DOI] [PMC free article] [PubMed] [Google Scholar]