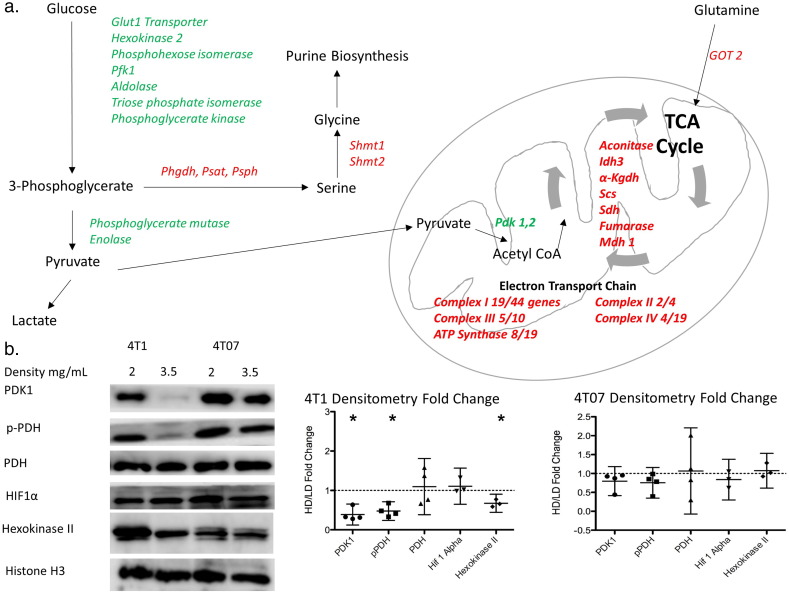
Fig. 4.
Microarray analysis reveals alterations in the expression of major pathways of cellular metabolism in response to changes in collagen extracellular matrix density.
A. Diagram of different enzymes in glycolysis, tricarboxylic acid cycle, electron transport chain and serine synthesis pathway regulated by changes in extracellular collagen matrix density in 4T1 cells. Green indicates downregulation in 4T1 cells in a HD collagen matrix while red indicates upregulation of gene in a HD collagen matrix. (N = 5, Fold change >± 1.2, and filtered for significance p < 0.01). For Complex I to IV, the number of genes altered in the complex is expressed as a fraction of the total number of genes that contribute to that complex. B. Representative images for protein immunoblotting for some enzymes in each metabolic pathway. C. Densitometry changes for these immunoblots in 4T1 and 4T07 cells. (HD/LD mean fold change ± 95% confidence interval) (4T1 HD/LD confidence interval for PDK1 = 0.130–0.697, pPDH = 0.241–0.752, Hexokinase II = 0.539–0.809. p < 0/05 by one-sample t-test.).
(PFK1 – Phosphofructokinase 1, PHGDH – 3-Phosphoglycerate dehydrogenase, PSAT – Phosphoserine aminotransferase, PSPH – Phosphoserine phosphatase, SHMT – Serine hydroxymethyl transferase, PDK – Pyruvate dehydrogenase kinase, IDH – Isocitrate dehydrogenase, α-KGDH – α-ketoglutarate dehydrogenase, SCS – Succinyl coA synthetase, SDH – Succinate dehydrogenase, MDH – Malate dehydrogenase, GOT – Glutamate oxaloacetate transaminase, PDH – Pyruvate dehydrogenase, HIF – Hypoxia inducible factor).