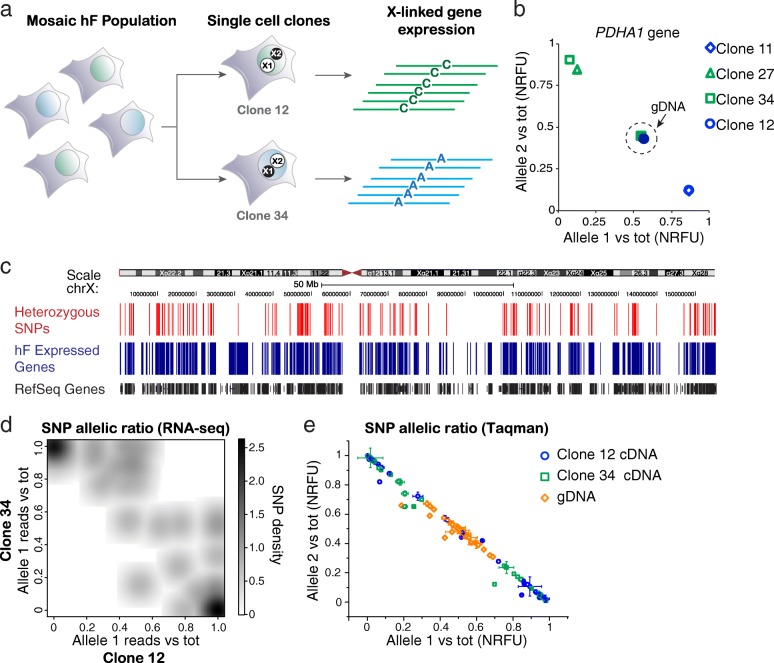
Fig. 1.
Allele-specific expression in single cell-derived clones discriminates the Xa and Xi. a Scheme illustrates the experimental design. Single cells that express/silence reciprocal X chromosomes (i.e. X1aX2i or X1iX2a) were isolated from a female (X1X2) hF line with balanced XCI (i.e. NHDF17914) and expanded to derive reciprocal isogenic clones (e.g. clone 12 and 34). Expression analysis of heterozygous SNPs within X-linked genes then allowed the active (Xa, open white circle) and inactive (Xi, closed black circle) X chromosomes in each clone to be distinguished. b Plot showing allele-specific expression of the X-linked gene PDHA1 in four hF clones where the allelic ratio was determined by SNP-specific Taqman probes and reported as normalised relative fluorescent units (NRFU) vs. total (allele1 + allele2). Using a known heterozygous SNP (rs1042456), clones 11 and 12 (blue diamond and circle) are shown to express a distinct allele from clones 27 and 34 (green triangle and square) while both alleles were detected in genomic (gDNA) hF samples (dashed circle). c Heterozygous SNPs identified by RNA-seq (i.e. 379 SNPs by strategy 1 in Additional file 1: Figure S1) are represented as red lines along the X-chromosome ideogram. Genes expressed in fibroblasts (FPKM >25th percentile) and reference genes (USCS Genome browser) are shown in blue and black, respectively. d Density plot represents the overall allelic expression in clone 12 and 34 obtained by RNA-seq ahead of data modelling. For each of the 379 heterozygous SNPs, the ratio between the SNP-reads overlapping allele 1 (i.e. the ones matching the SNP reported in the Reference genome sequence) vs. the total is shown, where the intensity of grey scale represents the density of SNPs with a certain allelic ratio. e Plot shows allelic ratios of 52 heterozygous SNPs that were identified by RNA-seq. Data represent the average of at least two independent samples for each clone and are plotted as normalised relative fluorescent units (NRFU) of each allele vs. the total. Error bars indicate standard errors of mean (SEM)