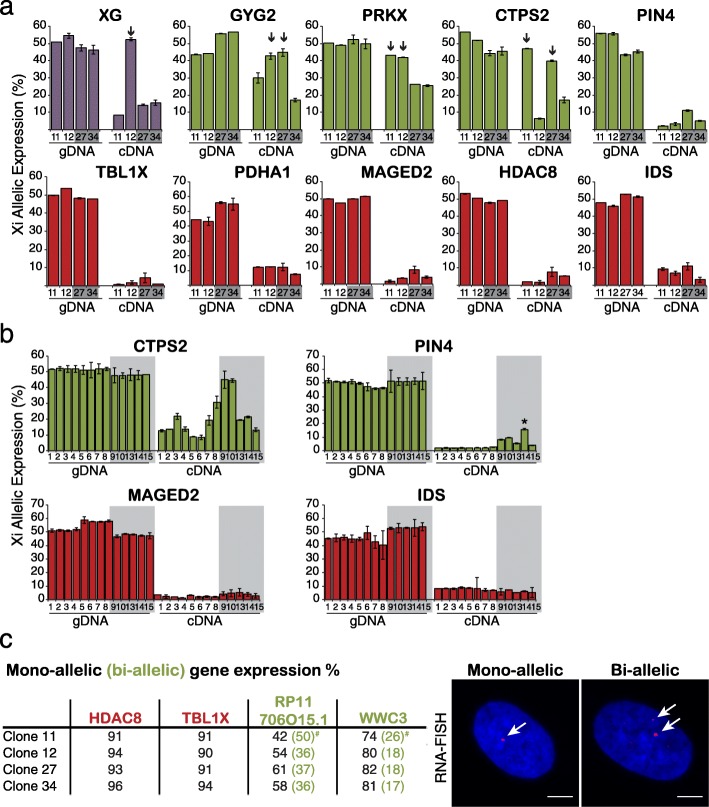
Fig. 5.
Variable allelic expression of Xi genes in hFs predicts their susceptibility to reprograming-mediated reactivation. a Histograms show Xi-allelic expression (as a percentage of total) of 10 X-linked genes in hF clones 11 and 12 and reciprocal clones 27 and 34 (grey), where gDNA provides controls. Allelic expression was measured using Taqman probes as NRFU of the Xi-specific probe vs. the total; data for each hF clone represent the average of at least three independent cultures. Arrows indicate significant differences in Xi expression (p <0.05, two-sided t-test) as compared with data from other clones. Genes that accordingly to RNA-seq are stably expressed (purple), reactivated (green) or remain inactive (red) across reprograming in clones 12 or 34 are shown. b Xi-allelic expression of CTPS2, PIN4 (reactivated genes), MAGED2 and IDS (inactive) in 13 additional hF clones. Analysis was performed as in (a). Asterisk (*) marks clones with Xi expression significantly different from the others (p <0.05, Grubbs test). c Mono-allelic and bi-allelic expression of X-linked genes in hFs was determined using RNA-FISH (exemplified in the images shown to the right) where punctate signals (red, arrowed) represent transcribed X-linked gene loci. At least 200 nuclei containing either a single punctate signal (mono-allelic) or two separate signals (bi-allelic) were scored for each hF clone and the percentage of total cells showing mono-allelic or bi-allelic (in brackets) patterns is shown. For HDAC8 and TBL1X >90% of the analysed cells showed mono-allelic expression. # indicates samples with higher bi-allelic expression than others (RNA-FISH) where significant Xi expression was also detected (RNA-seq). DAPI (blue) stains nuclei and scale bars indicate 5 μm