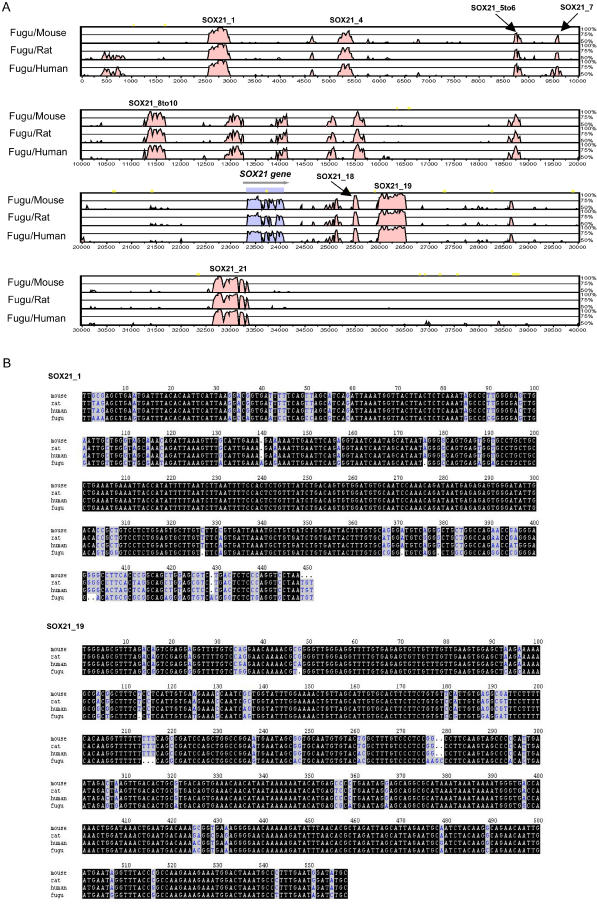
Figure 3. Comparative Sequence Analysis of the SOX21 Gene.
SOX21 genomic regions for mouse, human, and rat were extracted from Ensembl to include all flanking DNA up to the nearest neighbouring genes (ABCC4 and NM_180989 in the human genome and their orthologues in the rodent genomes). The region covering Fugu SOX21 (138–178 kb of Fugu Scaffold_293 [M000293]) was extracted from the Fugu Genome Server at http://fugu.rfcgr.mrc.ac.uk/fugu-bin/clonesearch.
(A) MLAGAN alignment of the SOX21 gene using Fugu DNA as the base sequence compared with mouse, rat, and human genomic DNA. Coloured peaks represent regions of sequence conservation above 60% over at least 40 bp. The SOX21 coding region (SOX21 is a single exon gene) is annotated, and sequence identity is shaded in blue. Non-coding regions of sequence identity are shaded in pink. The eight elements that have been functionally assayed are labelled. Six of these are identified in the global analysis as seven CNEs (SOX21_8–10 covers two CNEs). SOX21_7 and SOX21_18 are rCNEs.
(B) Multiple DNA sequence alignments of CNE SOX21_1 and CNE SOX21_19 between mouse, rat, human, and Fugu.