Abstract
The serrated neoplasia pathway of colorectal carcinogenesis is characterized by BRAF mutation and aberrant DNA methylation, which have not been reported on Korean patients. The aim of this study was to investigate BRAF mutation and DNA methylation in colorectal serrated polyps and the right colon.
Between 2005 and 2013, 146 colon polyps (47 tubular adenomas [TAs], 53 traditional serrated adenomas [TSAs], 17 sessile serrated adenomas/polyps [SSAs], and 29 hyperplastic polyps in the proximal colon [PHPs]) were collected from patients. Paraffin-embedded colon polyp tissue was used for DNA extraction. BRAF V600E mutation was identified through polymerase chain reaction (PCR) and pyrosequencing assay. The methylation status of the long interspersed nucleotide element-1, insulin-like growth factor binding protein 7 (IGFBP7), mutL homolog 1 (hMLH1), and CD133 genes were evaluated through disulfite conversion, PCR, and pyrosequencing assay.
BRAF V600E mutation was found in 2.1% of TAs, 47.2% of TSAs, 41.2% of SSAs, and 20.7% of PHPs. TSA and SSA had higher BRAF mutation rates than did TA (P < 0.0001). TSA had higher BRAF mutation rates than did PHP (P = 0.018). IGFBP7 hypermethylation was found in 17% of TAs, 37.7% of TSAs, 88.2% of SSAs, and 37.5% of PHPs. TSA and SSA had higher hypermethylation of IGFBP7 than did TA (P = 0.021 and P < 0.0001, respectively). SSA had higher hypermethylation of IGFBP7 than did PHP (P = 0.002). hMLH1 hypermethylation was found in 2.1% of TAs, 5.7% of TSAs, 0% of SSAs, and 0% of PHPs. CD133 hypermethylation was found in 21.3% of TAs, 9.4% of TSAs, 35.3% of SSAs, and 17.4% of PHPs.
BRAF mutation and methylation in TSA and SSA are different from those in PHP in Koreans. These findings suggested that PHP may have different molecular characteristics compared with other serrated polyps.
Keywords: hyperplastic polyp, sessile serrated adenoma, traditional serrated adenoma
1. Introduction
The worldwide incidence of colorectal cancer (CRC) consists of about 1.2 million new cases per year; its mortality is relatively high, with as many as 40% to 50% of CRC patients dying within 5 years of diagnosis.[1–3] Three pathways of CRC carcinogenesis are as follows: the chromosomal instability pathway, mutator pathway, and serrated neoplasia pathway.[4–6] The serrated neoplasia pathway suggests that serrated polyps having saw-toothed involution of the crypt epithelium may develop into CRC with gene promoter hypermethylation and microsatellite instability.[7,8] Thirty years ago, serrated lesions in the colorectum were named hyperplastic polyps, and were considered to have no malignant potential. However, serrated polyps have since been revealed to be the precursors of approximately one-third of CRCs.[9–11]
Recent studies have shown that the serrated neoplasia pathway, characterized by BRAF mutation and CpG island methylator phenotype (CIMP), with or without microsatellite instability, is involved in CRC development from serrated polyps. Currently, serrated lesions are classified into hyperplastic polyps, sessile serrated adenoma/polyp with or without cytological dysplasia, and traditional serrated adenoma, according to World Health Organization (WHO) criteria.[11] The 3 types of serrated lesions, individually or in combination, can be involved in the serial transition from normal epithelium to polyps, and to CRC.[1]
BRAF is a member of the RAF family of serine/threonine kinases that mediates cellular responses to growth signals through the RAS-RAF-mitogen-activated protein kinase pathway. BRAF mutations play a role in the chromosomal instability pathway of CRC and have recently been found in 5% to 15% of CRCs.[12] Methylation of CpG islands within promoter regions of genes is a normal mechanism of gene expression reduction. When a tumor-suppressor gene is methylated, the reduced expression may cause carcinogenesis. The extent of hypermethylation of promoter CpG islands in neoplasms varies considerably.[11] The microsatellite instability (MSI) pathway of CRC is characterized by the loss of mismatch repair gene function, which leads to DNA replication errors. Loss of mismatch repair usually occurs because of germline mutation of 1 of 4 mismatch-repair genes (MSH2, MLH1, MSH6, or PMS2). Recent studies have suggested that methylation of insulin-like growth factor binding protein 7 (IGFBP7) is an important alteration in the serrated neoplasia pathway and correlates with MLH1 methylation, BRAF mutation, CIMP, and MSI in CRC. However, the pathological and epigenetic features of serrated polyps with methylated IGFBP7 are still largely unknown.[13] Long interspersed nucleotide element-1 (LINE-1) is a global DNA methylation marker,[14] whose hypomethylation predicts a poor outcome in several types of human neoplasms, such as colon, stomach, and ovarian cancer.[15,16] CD133 is a cancer stem cell marker of CRC.[17]
The aim of this study was to investigate BRAF mutation and DNA methylation in serrated polyps of the colorectum in Koreans.
2. Materials and methods
2.1. Tissues
Colorectal polyps removed during colonoscopy in the Gastrointestinal Endoscopy Center, Wonju Severance Christian Hospital, Wonju, Republic of Korea, from 2005 to 2013, were collected for analysis. The colorectal polyps were classified into 4 groups: tubular adenoma (TA), traditional serrated adenoma (TSA), sessile serrated adenoma/polyp (SSA), and hyperplastic polyp in the proximal colon (PHP), by an expert pathologist (MYC). All polyps of TSA, SSA, and PHP were removed between 2005 and 2013, and stored as paraffin blocks. TA samples were selected randomly from all TA polyps. In TSA, those of size greater than 10 mm were selected. The polyps had been prepared as formalin-fixed, paraffin-embedded (FFPE) tissue blocks. All tissues were reviewed by an experienced pathologist (MYC). Ultimately, a total of 146 colorectal polyps were included in this study, including 53 cases of TSA, 47 cases of TA, 29 cases of PHP, and 17 cases of SSA. Of the 17 SSAs, 2 cases had low-grade dysplasia and 15 cases had no cytological dysplasia. This study was approved by the Institutional Ethics Committee of Yonsei University, Wonju Severance Christian Hospital (CR312041), Wonju, Republic of Korea.
2.2. DNA extraction and polymerase chain reaction analysis
2.2.1. BRAF V600E mutation
The paraffin-embedded tissues were prepared as 5-μm-thick histological sections, stained with hematoxylin and eosin, and the pathologic foci were microdissected using a surgical scalpel under microscopic guidance. The microdissected tissues were deparaffinized with xylene, and subjected to proteinase K digestion at 56°C for 1 hour. Genomic DNA was extracted from the deparaffinized tissues using the QIAamp DNA FFPE Tissue Kit and the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany). The primers for polymerase chain reaction (PCR) of BRAF V600E mutation were as follows: forward, 5’-GAAGACCTCACAGTAAAAATAG-3’ and reverse, 5’-ATAGCCTCAATTCTTACCATCC-3’ (Table 1).[18] The PCR conditions included initial denaturation at 95°C for 10 minutes, followed by 45 cycles of denaturation at 95°C for 30 seconds, annealing at 51 to 60°C for 30 seconds, and elongation at 72°C for 30 seconds. The reaction ended with a final extension at 72°C for 5 minutes. The PCR products were analyzed by electrophoresis in a 2% agarose gel to confirm successful amplification of PCR product.
Table 1.
Primer sequences and conditions.

2.2.2. Methylation of LINE-1, hMLH1, IGFBP7, and CD133
The paraffin-embedded tissues were prepared as 5-μm-thick histological sections, stained with hematoxylin and eosin, and the pathologic foci were microdissected using a surgical scalpel under microscopic guidance. The microdissected tissues were deparaffinized with xylene, and subjected to proteinase K digestion at 56°C for 1 hour. Genomic DNA was extracted from the deparaffinized tissues using the QIAamp DNA FFPE Tissue Kit and the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany), and treated with bisulfite using the EpiTect Bisulfite Kit (Qiagen, Hilden, Germany) to convert unmethylated cytosines to uracils according to the manufacturer's protocols. The primers for PCR of LINE-1, IGFBP7, mutL homolog 1 (hMLH1), and CD133 were as follows: LINE-1 forward (5’-TTTTGAGTTAGGTGTGGGATATA-3’) and reverse (5’-biotin-AAAATCAAAAAATTCCCTTTC-3’), IGFBP7 forward (5’-AGGGTTYGGGGTAGGGGATTGGGGAT-3’) and reverse (5’-biotin-AAAACCACACCCCYAAACYATAAAAACAC-3’), hMLH1 forward (5’-TTGGTATTTAAGTTGTTTAATTAATAGTTG-3’) and reverse (5’-biotin-AAAATACCTTCAACCAATCACCTC-3’), and CD133 forward (5’-GAGTAGGGATATGGGGGTATAA-3’) and reverse (5’-biotin-AACACCCCAATTCTCCAT-3’) (Table 1). The PCR conditions included initial denaturation at 95°C for 10 minutes, followed by 45 cycles of denaturation at 95°C for 30 seconds, annealing at 51 to 60°C for 30 seconds, and elongation at 72°C for 30 seconds. The reaction ended with a final extension at 72°C for 5 minutes. The PCR products were analyzed by electrophoresis in a 2% agarose gel to confirm successful amplification of PCR product.
2.3. Pyrosequencing assay
Pyrosequencing was carried out using the PSQ96MA system (Biotage, Uppsala, Sweden), Pyro Q-CpG software (Biotage, Uppsala, Sweden), and a PyroMark kit (Qiagen, Hilden, Germany). PCR product was bound to Streptavidin-Sepharose HP (GE Healthcare, Uppsala, Sweden), purified, washed, denatured using a 0.2 M NaOH solution, and washed again. Strand separation and purification of the single-stranded template were facilitated using the Vacuum Prep Tool (Biotage, Uppsala, Sweden). The purified single-stranded template was then added to the sequencing mixture containing annealing buffer and the following sequencing primers: BRAF: 5’-GATTTTGGTCTAGCTACA-3’, LINE-1: 5’-AGTTAGGTGTGGGATATAGT-3’, IGFBP7: YGGGTGTTYGTTTATTTT-3’, hMLH1: 5’- AGTTATAGTTGAAGGAAGAA -3’, and CD133: 5’- GGGATATGGGGGTATAAAG -3’, as per published reports (Table 1),[19–21] then incubated at 85°C for 2 minutes, and allowed to cool to room temperature. Pyrosequencing was carried out in a PyroMark ID instrument (Biotage, Uppsala, Sweden), and the data were analyzed using sequence analysis software from the manufacturer. The amount of C relative to the sum of the amounts of C and T at each CpG site was calculated as a percentage. The average of the relative amounts of C in the CpG sites was defined as the overall methylation level of each gene in the colon polyps.
2.4. Definition
The proximal colon was defined as the cecum, ascending colon, and transverse colon. The distal colon was defined as the descending colon, sigmoid colon, and the rectum. For LINE-1, if the percent of methylated reference (PMR) was ≥60, LINE-1 was defined as hypermethylated, and if the PMR was <60, LINE-1 was defined as hypomethylated. For IGFBP7, PMR ≥20 was defined as hypermethylated and PMR <20 was defined as hypomethylated. For hMLH1 and CD133, PMR ≥10 was defined as hypermethylated and PMR <10 was defined as hypomethylated.
2.5. Statistical analysis
All statistical analyses were performed using PASW (version 20.0) (SPSS Inc., Chicago, IL). Differences between groups were evaluated using 1-way analysis of variance (ANOVA) with Scheffe, chi-square test, and Fisher exact tests. Statistical significance tests were 2-tailed, and P < 0.05 was considered statistically significant.
3. Results
3.1. Baseline characteristics
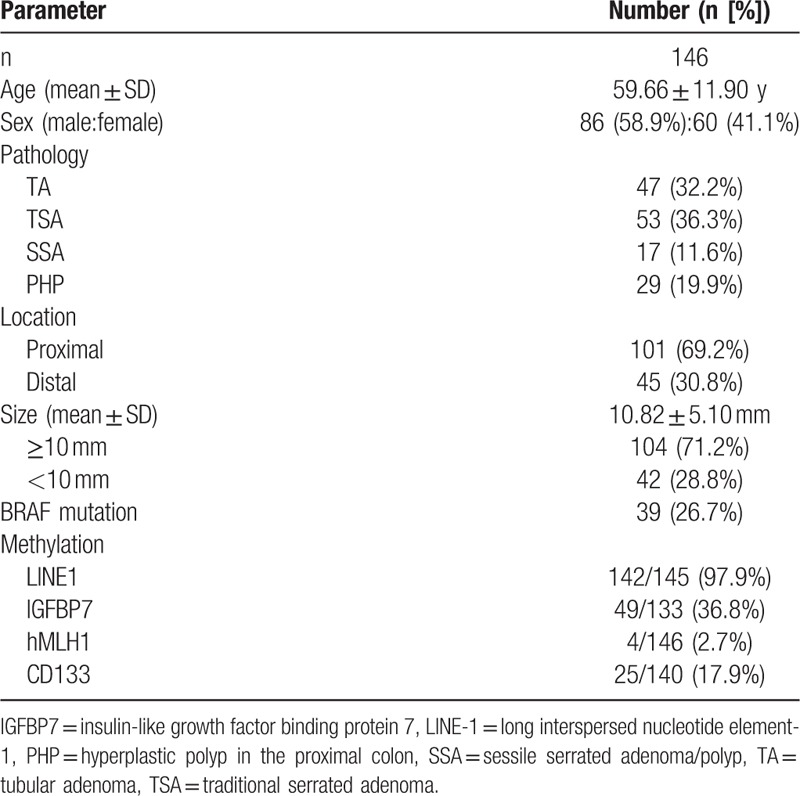
A total of 146 polyps were derived from 146 patients with a mean age of 59.66 ± 11.90 years (Table 2). They consisted of 86 (58.9%) males and 60 (41.1%) females. Of the 146 polyps, 101 (69.2%) polyps were located in the proximal colon and 45 (30.8%) polyps were located in the distal colon. The mean size of the 146 polyps was 10.82 ± 5.10 mm, and 104 (71.2%) polyps were ≥10 mm in size, whereas 42 (28.8%) polyps were <10 mm in size. BRAF V600E mutation was found in 39 (26.7%) polyps. The rates of hypermethylation in LINE-1, IGFBP7, hMLH1, and CD133 were 97.9% (142/145), 36.8% (49/133), 2.7% (4/146), and 17.9% (25/140), respectively.
Table 2.
Clinical features of all patients.
3.2. Clinical features of the 4 types of colon polyps
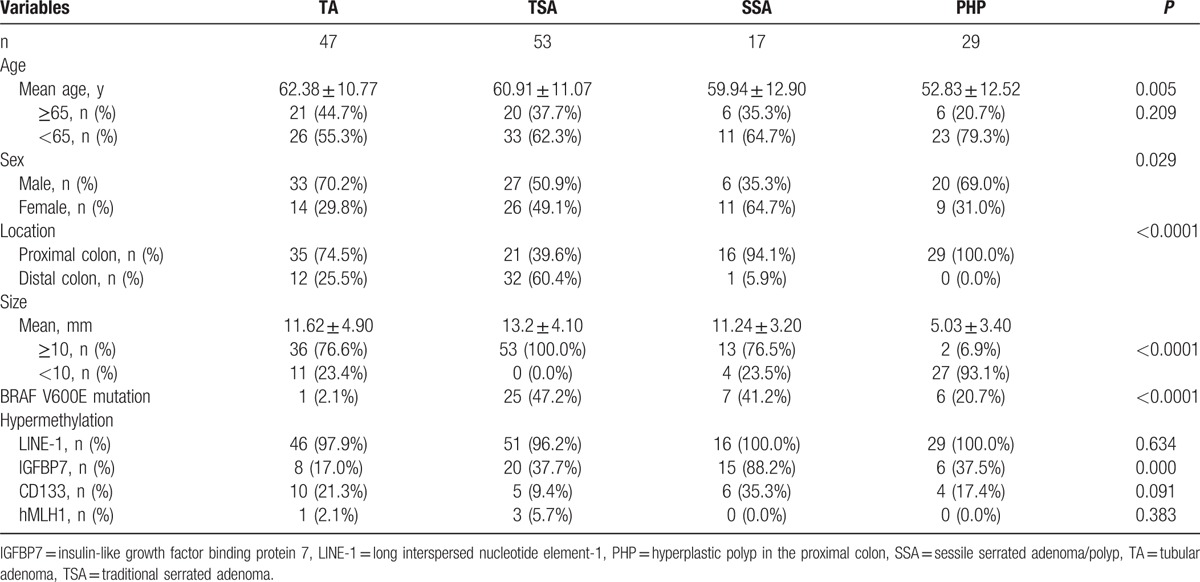
Among the 4 groups, patients with PHP had the lowest mean age (P = 0.005) (Table 3). Among patients with TA, TSA, and SSA, there were no significant differences in mean age. Patients with SSA had a lower percentage of male sex than did those with TA (70.2% vs 35.5%; P = 0.011). TA and SSA were mainly located in the proximal colon (74.5% and 94.1%, respectively). TSA was located in the distal colon more commonly than was TA (60.4% vs 25.5%; P < 0.0001). The mean polyp sizes were 11.62 ± 4.90 mm in TA, 13.2 ± 4.10 mm in TSA, 11.24 ± 3.20 mm in SSA, and 5.03 ± 3.40 mm in PHP.
Table 3.
Clinical features, BRAF mutation, and hypermethylation of LINE-1, IGFBP7, hMLH1, and CD133 in the 4 types of colon polyps.
3.3. BRAF V600E mutation
Traditional serrated adenoma and SSA had higher rates of BRAF V600E mutation than did TA (47.2% vs 2.1%; P < 0.0001 and 41.2% vs 2.1%; P < 0.0001, respectively) (Table 3). TSA had a higher rate of BRAF V600E mutation than did PHP (47.2% vs 20.7%; P = 0.018), but SSA was not significantly different in its rate of BRAF V600E mutation from PHP (41.2% vs 20.7%; P = 0.136) (Table 3).
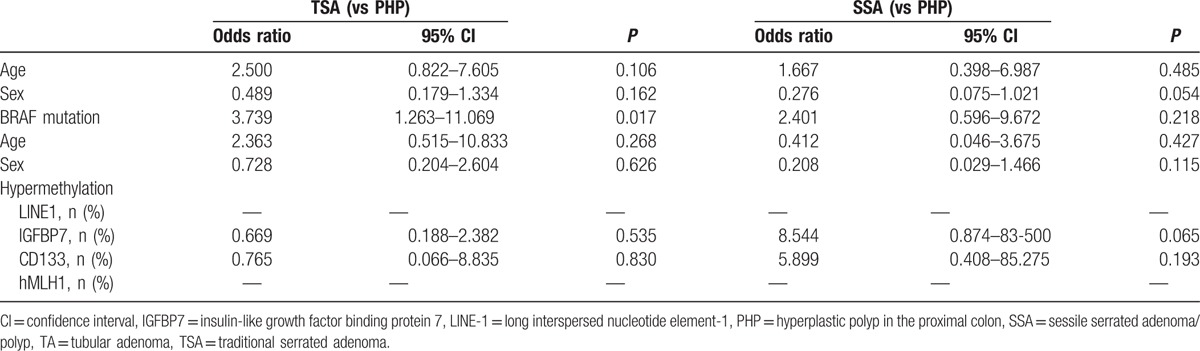
In multivariate analysis, BRAF V600E mutation was significantly associated with TSA and SSA, with TA as the reference (odds ratio [OR] 86.313, 95% confidence interval [CI] 7.573–983.727, P < 0.0001; and OR 28.436, 95% CI 2.578–313.632, P = 0.006) (Table 4). BRAF V600E mutation was significantly associated with TSA, with PHP as the reference (OR 3.739, 95% CI 1.263–11.069, P = 0.017), but not associated with SSA, with PHP as the reference (Table 5).
Table 4.
Multivariable analysis for risk factors of TSA and SSA, as TA was the reference.
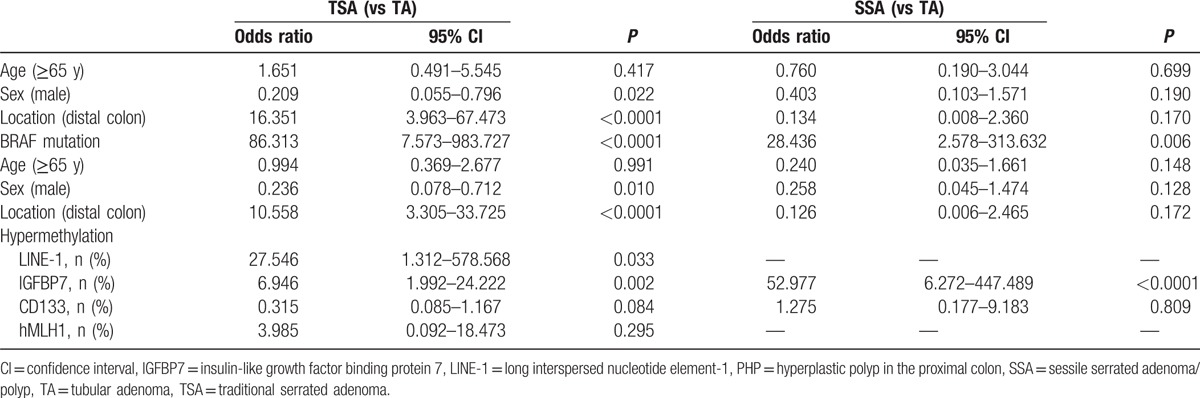
Table 5.
Multivariable analysis for risk factors of TSA and SSA, as PHP was the reference.
3.4. Hypermethylation of LINE-1, IGFBP7, hMLH1, and CD133
In univariate analysis, TSA had a lower hypermethylation rate of LINE-1 than did TA or SSA (P = 0.008 and P < 0.0001, respectively) (Table 3). PHP had a higher hypermethylation rate of LINE-1 than did TSA (P < 0.0001), but PHP did not vary from SSA in its hypermethylation rate of LINE-1 (P = 0.471). With respect to the hypermethylation rate of IGFBP7, TSA and SSA were higher than TA (P = 0.021 and P < 0.0001, respectively), and SSA was higher than TSA (P < 0.001). PHP had a lower rate than did SSA (P = 0.002). The hypermethylation rate of hMLH1 was very low in all 4 types. The hypermethylation rates of CD133 were not significantly different among the 4 types.
In multivariable analysis, TSA exhibited a significant association with hypermethylation of LINE-1 (OR 27.546, 95% CI 1.312–578.568, P = 0.033) and of IGFBP7 (OR 6.946, 95% CI 1.992–24.222, P = 0.002), with TA as the reference (Table 4). SSA was associated with hypermethylation of IGFBP7 (OR 52.977, 95% CI 6.272–447.489, P < 0.0001), with TA as the reference (Table 4). With TA as the reference, TSA and SSA were not associated with hypermethylation of CD133 (Table 4).
4. Discussion
Our study showed that TSA and SSA had a higher rate of BRAF V600E mutation than did TA, and TSA had a higher rate of BRAF V600E mutation than did PHP.
BRAF V600E mutation is rare in tubular adenomas of the colon, whereas BRAF V600E mutation is frequent in serrated polyps.[22–25] The rate of BRAF mutation has been reported to be 62.1% to 90% in SSA, and 27% to 55% in TSA.[22–25] One Chinese study reported a 14.3% BRAF V600E mutation rate in SSA, which is a lower rate than in the Western population.[26] One Korean study reported that BRAF V600E mutations are found in 43.5% to 58.3% of TSAs.[27] In our study, the rates of BRAF V600E mutation were 47.2% in TSA and 41.2% in SSA, consistent with other studies. BRAF V600E mutation is an early event in the serrated neoplasia pathway.[2] BRAF V600E mutation has been suggested to be a specific marker of both serrated polyps and the serrated neoplasia pathway.[23] Specifically, proximal serrated polyps with BRAF V600E mutation have a high risk of progression to malignancy.[28] The mitogen-activated protein kinase (MAPK) signaling pathway is generally altered in CRC and precursor lesions by oncogenic mutation of either the BRAF or KRAS genes. These mutations are mutually exclusive and revealed a striking specificity for the serrated polyp subtype.[29] In our study, TSA and SSA had significantly higher rates of BRAF V600E mutation than did tubular adenoma. PHP had more BRAF mutation than TSA. Although these findings did not have clinical implication, those suggested that PHP maybe have different molecular characteristics compared with serrated polyps.
Insulin-like growth factor binding protein 7 is expressed in various tissues, such as the lung, brain, prostate, and gastrointestinal tract. It is inactivated by DNA methylation in the human colon, and has a potential tumor-suppressor role against colorectal carcinogenesis.[30] Methylation of IGFBP7 may play a key role in the serrated neoplasia pathway, and IGFBP7 methylation occurs during the course from serrated polyp to cancer with MSI.[13] One study by Kaji et al[13] suggested that methylation of IGFBP7 is associated with the formation of a saw-tooth architecture, and that inactivation of IGFBP7 is an early event in colorectal tumorigenesis. In our study, IGFBP7 was highly methylated in SSA (88.2%) compared with that in TA (17.0%), TSA (37.7%), or PHP (37.5%). An Australian study reported that the rates of IGFBP7 methylation are 62.9% in SSA and 32.1% in hyperplastic polyps.[28]
Long interspersed nucleotide element-1 is an indicator of genome-wide DNA methylation level. Genome-wide DNA hypomethylation plays an important role in genomic instability and carcinogenesis.[14] LINE-1 methylation is inversely associated with MSI and CpG island methylator phenotype. LINE-1 hypomethylation is associated with poor prognosis in CRC.[31] One Japanese study reported that TSA with high grade dysplasia had high levels of methylation of LINE-1, and concluded that hypomethylation of LINE-1 may have a potential role in TSA pathway progression.[32] In our study, LINE1 was highly methylated in TA (97.9%), TSA (96.2%), SSA (100.0%), and PHP (100.0%).
Methylation of CD133 is rare in serrated lesions and polyps.[33] In our study, methylation rates of CD133 were found to be 35.3% in SSA and 9.4% in TSA. Methylation of CD133 is a late alteration leading to dysplasia in CRCs.[33,34]
In our study, hMLH1 methylation rates were very low. hMLH1 methylation is observed in the proximal colon and is associated with BRAF mutation.[28] One study reported that hMLH1 methylation was found in 7% of tubular adenomas and 7% of hyperplastic polyps.[33] Another study reported that the dysplastic component of sessile serrated lesions had higher methylation levels of hMLH1 than did the nondysplastic component (12.94% vs 5.89%).[35] Methylation of hMLH1 is a late alteration leading to dysplasia in CRCs.[33,34]
This study had several limitations. The study population was collected from a single center, and did not represent the general Korean population. Thus selection bias cannot be excluded. However, to the best of our knowledge, this is the first study to investigate the molecular characteristics of TSA, SSA, and PHP in Koreans. The sample size was not sufficient to fully conclude the methylation status of serrated lesions. In addition, the tubular adenomas were selected randomly, but not matched, and should be matched by sex and age to SSA or TSA in future studies. CpG island methylator phenotype analysis and microsatellite instability was not examined because extracted DNA was not enough. Instead, CD133 was used as a global DNA methylation marker. In addition, CD133 is a marker of the cancer stem cell population, and methylation of cancer stem cell-related Wnt target genes including CD133 may reflect poor prognosis in CRC.[36]
In conclusion, this study showed that BRAF V600E mutation was more frequently found in TSA and SSA than in PHP. PHP had different methylation profile compared with TSA and SSA. These findings suggest that PHP may have different molecular characteristics compared with serrated polyps, although their clinical implication are uncertain.
Footnotes
Abbreviations: CIMP = CpG island methylator phenotype, CRC = colorectal cancer, FFPE = formalin-fixed, paraffin-embedded, IGFBP7 = insulin-like growth factor binding protein 7, LINE-1 = long interspersed nucleotide element-1, PCR = polymerase chain reaction, PHP = hyperplastic polyp in the proximal colon, SSA = sessile serrated adenoma/polyp, TA = tubular adenoma, TSA = traditional serrated adenoma, WHO = World Health Organization.
OS and HMK are the first 2 authors and contributed equally to this study.
Funding: This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2012–0004734) and the grant-in aid Yonsei University Wonju College of Medicine.
The authors have no conflict of interests.
References
- [1].Gaiser T, Meinhardt S, Hirsch D, et al. Molecular patterns in the evolution of serrated lesion of the colorectum. Int J Cancer 2013;132:1800–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mesteri I, Bayer G, Meyer J, et al. Improved molecular classification of serrated lesions of the colon by immunohistochemical detection of BRAF V600E. Mod Pathol 2014;27:135–44. [DOI] [PubMed] [Google Scholar]
- [3].Zhang NH, Li J, Li Y, et al. Co-expression of CXCR4 and CD133 proteins is associated with poor prognosis in stage II-III colon cancer patients. Exp Ther Med 2012;3:973–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Winawer SJ, Zauber AG, Ho MN, et al. Prevention of colorectal cancer by colonoscopic polypectomy. The National Polyp Study Workgroup. N Engl J Med 1993;329:1977–81. [DOI] [PubMed] [Google Scholar]
- [5].Brenner H, Chang-Claude J, Seiler CM, et al. Case-control study supports extension of surveillance interval after colonoscopic polypectomy to at least 5 yr. Am J Gastroenterol 2007;102:1739–44. [DOI] [PubMed] [Google Scholar]
- [6].Lieberman DA, Weiss DG, Harford WV, et al. Five-year colon surveillance after screening colonoscopy. Gastroenterology 2007;133:1077–85. [DOI] [PubMed] [Google Scholar]
- [7].Leggett B, Whitehall V. Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology 2010;138:2088–100. [DOI] [PubMed] [Google Scholar]
- [8].O’Brien MJ, Yang S, Mack C, et al. Comparison of microsatellite instability, CpG island methylation phenotype, BRAF and KRAS status in serrated polyps and traditional adenomas indicates separate pathways to distinct colorectal carcinoma end points. Am J Surg Pathol 2006;30:1491–501. [DOI] [PubMed] [Google Scholar]
- [9].Lane N. The precursor tissue of ordinary large bowel cancer. Cancer Res 1976;36:2669–72. [PubMed] [Google Scholar]
- [10].Sumner HW, Wasserman NF, McClain CJ. Giant hyperplastic polyposis of the colon. Dig Dis Sci 1981;26:85–9. [DOI] [PubMed] [Google Scholar]
- [11].Rex DK, Ahnen DJ, Baron JA, et al. Serrated lesions of the colorectum: review and recommendations from an expert panel. Am J Gastroenterol 2012;107:1315–29. [quiz 1314, 1330]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chan TL, Zhao W, Leung SY, et al. BRAF and KRAS mutations in colorectal hyperplastic polyps and serrated adenomas. Cancer Res 2003;63:4878–81. [PubMed] [Google Scholar]
- [13].Kaji E, Uraoka T, Kato J, et al. Externalization of saw-tooth architecture in small serrated polyps implies the presence of methylation of IGFBP7. Dig Dis Sci 2012;57:1261–70. [DOI] [PubMed] [Google Scholar]
- [14].Suzuki K, Suzuki I, Leodolter A, et al. Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell 2006;9:199–207. [DOI] [PubMed] [Google Scholar]
- [15].Shigaki H, Baba Y, Watanabe M, et al. LINE-1 hypomethylation in gastric cancer, detected by bisulfite pyrosequencing, is associated with poor prognosis. Gastric Cancer 2013;16:480–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Goodier JL, Cheung LE, Kazazian HH., Jr Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition. Nucleic Acids Res 2013;41:7401–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Horst D, Kriegl L, Engel J, et al. Prognostic significance of the cancer stem cell markers CD133, CD44, and CD166 in colorectal cancer. Cancer Invest 2009;27:844–50. [DOI] [PubMed] [Google Scholar]
- [18].Jo YS, Huang S, Kim YJ, et al. Diagnostic value of pyrosequencing for the BRAF V600E mutation in ultrasound-guided fine-needle aspiration biopsy samples of thyroid incidentalomas. Clin Endocrinol (Oxf) 2009;70:139–44. [DOI] [PubMed] [Google Scholar]
- [19].Jeon YK, Kim SH, Choi SH, et al. Promoter hypermethylation and loss of CD133 gene expression in colorectal cancers. World J Gastroenterol 2010;16:3153–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kimura T, Yamamoto E, Yamano HO, et al. A novel pit pattern identifies the precursor of colorectal cancer derived from sessile serrated adenoma. Am J Gastroenterol 2012;107:460–9. [DOI] [PubMed] [Google Scholar]
- [21].Dammann RH, Kirsch S, Schagdarsurengin U, et al. Frequent aberrant methylation of the imprinted IGF2/H19 locus and LINE1 hypomethylation in ovarian carcinoma. Int J Oncol 2010;36:171–9. [PubMed] [Google Scholar]
- [22].Jass JR, Baker K, Zlobec I, et al. Advanced colorectal polyps with the molecular and morphological features of serrated polyps and adenomas: concept of a ’fusion’ pathway to colorectal cancer. Histopathology 2006;49:121–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rosenberg DW, Yang S, Pleau DC, et al. Mutations in BRAF and KRAS differentially distinguish serrated versus non-serrated hyperplastic aberrant crypt foci in humans. Cancer Res 2007;67:3551–4. [DOI] [PubMed] [Google Scholar]
- [24].Mohammadi M, Kristensen MH, Nielsen HJ, et al. Qualities of sessile serrated adenoma/polyp/lesion and its borderline variant in the context of synchronous colorectal carcinoma. J Clin Pathol 2012;65:924–7. [DOI] [PubMed] [Google Scholar]
- [25].Fu B, Yachida S, Morgan R, et al. Clinicopathologic and genetic characterization of traditional serrated adenomas of the colon. Am J Clin Pathol 2012;138:356–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Qiu Y, Fu X, Zhang W, et al. Prevalence and molecular characterisation of the sessile serrated adenoma in a subset of the Chinese population. J Clin Pathol 2014;67:491–8. [DOI] [PubMed] [Google Scholar]
- [27].Kim MJ, Lee EJ, Suh JP, et al. Traditional serrated adenoma of the colorectum: clinicopathologic implications and endoscopic findings of the precursor lesions. Am J Clin Pathol 2013;140:898–911. [DOI] [PubMed] [Google Scholar]
- [28].Fernando WC, Miranda MS, Worthley DL, et al. The CIMP phenotype in BRAF mutant serrated polyps from a prospective colonoscopy patient cohort. Gastroenterol Res Pract 2014;2014:374926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rosty C, Hewett DG, Brown IS, et al. Serrated polyps of the large intestine: current understanding of diagnosis, pathogenesis, and clinical management. J Gastroenterol 2013;48:287–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Dimberg J, Hong TT, Skarstedt M, et al. Analysis of APC and IGFBP7 promoter gene methylation in Swedish and Vietnamese colorectal cancer patients. Oncol Lett 2013;5:25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ogino S, Nosho K, Kirkner GJ, et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst 2008;100:1734–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Naito T, Nosho K, Ito M, et al. IGF2 differentially methylated region hypomethylation in relation to pathological and molecular features of serrated lesions. World J Gastroenterol 2014;20:10050–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Petko Z, Ghiassi M, Shuber A, et al. Aberrantly methylated CDKN2A, MGMT, and MLH1 in colon polyps and in fecal DNA from patients with colorectal polyps. Clin Cancer Res 2005;11:1203–9. [PubMed] [Google Scholar]
- [34].Wynter CV, Walsh MD, Higuchi T, et al. Methylation patterns define two types of hyperplastic polyp associated with colorectal cancer. Gut 2004;53:573–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lee EJ, Chun SM, Kim MJ, et al. Reappraisal of hMLH1 promoter methylation and protein expression status in the serrated neoplasia pathway. Histopathology 2016;69:198–210. [DOI] [PubMed] [Google Scholar]
- [36].de Sousa E, Melo F, Colak S, et al. Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell 2011;9:476–85. [DOI] [PubMed] [Google Scholar]


