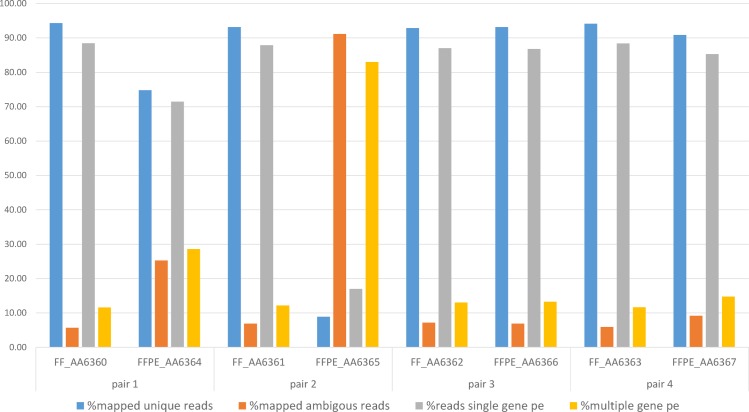
Fig 3. Mapped reads in FF and FFPE tissue samples.
Percentages of uniquely mapped paired-reads, ambiguously mapped paired-end reads, paired-end reads mapping into a single gene, and paired-end reads mapping into multiple genes. Note that the most degraded FFPE sample (AA_6365) had very high percentages of ambiguous reads (>90%) and reads mapping to multiple genes (>80%), whereas the second most degraded FFPE sample (AA_6364) had intermediate percentages (25% and ~30% respectively). The remaining samples had low percentages of ambiguities (~10%).