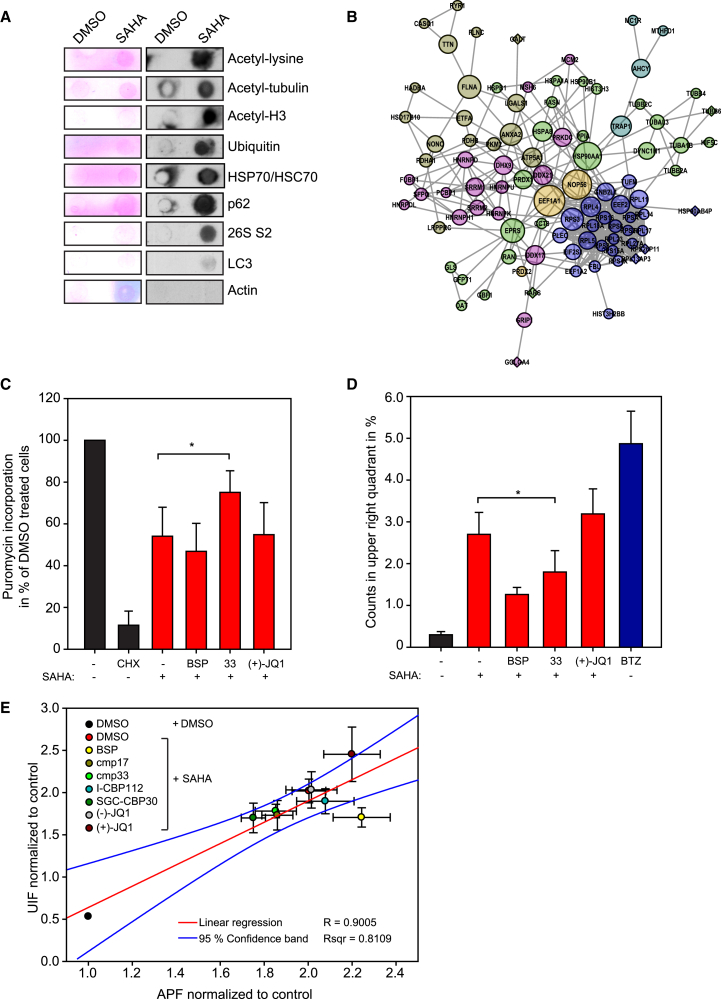
Figure 6.
Composition of the HDAC Inhibitor-Induced Aggregates and Effect of CBP/p300-Specific BDIs
(A) U2OS cells were treated with either DMSO or SAHA (5 μM) for 24 hr and then harvested; membranes and DNA were removed and the remaining aggregate fraction treated with detergent. The material captured in the filter retardation assay was visualized by Ponceau S staining or immunostaining with the indicated antibodies (see also Figures S6A and S6B).
(B) U2OS cells were treated with either DMSO or SAHA (5 μM) for 24 hr and then harvested; membranes and DNA were removed and the remaining aggregate fraction treated with detergent. The aggregated proteins were trypsinized and subjected to tandem mass spectrometry analysis. Proteins were grouped according to their biological role, which is symbolized by the colors. The size of the symbols represent the betweenness-centrality of the proteins, and the circles represent proteins known to be acetylated, either determined to be so in this study or by mining Phosida, UniProt, MaxQB, CPLM, and PhosphoSitePlus (see also Figure S6A).
(C) Cells were treated with SAHA (5 μM) and the indicated BDIs. The effect on mRNA translation was visualized by incorporation of puromycin in the nascent polypeptide chain after immunoblotting with a monoclonal antibody against puromycin in polypeptide chains. For comparison, Ponceau S served as a loading control. The intensities of the anti-puromycin signal in the different lanes were quantified and normalized against the intensities of the Ponceau S staining. The average of five independent experiments is shown, normalized to puromycin incorporation in percentage of DMSO (−)-treated cells. Level of statistical significance is indicated (*p ≤ 0.05). Error bars denote SD. Quantification of the independent biological replicates as shown in Figure S7A. BSP, bromosporine; 33, compound 33; (+)-J, (+)-JQ1.
(D) HEK293T cells stably expressing Ub-EGFP were treated with 5 μM of the HDAC inhibitor and with the indicated BDIs. Cells were then analyzed by flow cytometry; green fluorescence (FL1 channel) of the Ub-EGFP is plotted on the y axis and red fluorescence of Proteostat staining (FL3 channel) on the x axis, as shown in Figure S7B. Quadrants were chosen according to the DMSO (−) control, which is set to the lower left quadrant, and the FL3 mean fluorescence in the upper right quadrant of the gated cells was quantified. Shown are three biological replicates, each in technical triplicates. Level of statistical significance is indicated (*p ≤ 0.05). Error bars denote SD. BSP, bromosporine; 33, compound 33; (+)-J, (+)-JQ1.
(E) HEK293T cells stably expressing Ub-EGFP were treated with SAHA (5 μM) and the indicated BDIs (2.5 μM). Cells were then fixed, permeabilized, and stained with Proteostat. FACS was carried out and the cells were measured in the FL1 channel to detect accumulated Ub-EGFP and in the FL3 channel to detect Proteostat-positive aggregates. After gating and compensation, the mean fluorescence intensities in the upper right quadrants were plotted against each other. The data were derived from at least three independent biological replicates, each in technical triplicates. Linear regression resulted in R2 = 0.8109. BSP, bromosporine.
See also Figures S6 and S7.